|
Smith–Waterman Algorithm
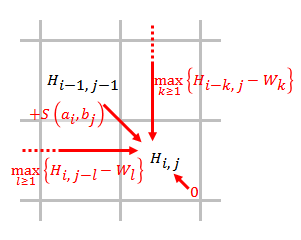
The Smith–Waterman algorithm performs local sequence alignment; that is, for determining similar regions between two strings of nucleic acid sequences or protein sequences. Instead of looking at the Needleman–Wunsch algorithm, entire sequence, the Smith–Waterman algorithm compares segments of all possible lengths and Mathematical optimization, optimizes the similarity measure. The algorithm was first proposed by Temple F. Smith and Michael S. Waterman in 1981. Like the Needleman–Wunsch algorithm, of which it is a variation, Smith–Waterman is a dynamic programming algorithm. As such, it has the desirable property that it is guaranteed to find the optimal local alignment with respect to the scoring system being used (which includes the substitution matrix and the Gap penalty, gap-scoring scheme). The main difference to the Needleman–Wunsch algorithm is that negative scoring matrix cells are set to zero, which renders the (thus positively scoring) local alignments visible ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are typically represented as rows within a matrix. Gaps are inserted between the residues so that identical or similar characters are aligned in successive columns. Sequence alignments are also used for non-biological sequences, such as calculating the distance cost between strings in a natural language or in financial data. Interpretation If two sequences in an alignment share a common ancestor, mismatches can be interpreted as point mutations and gaps as indels (that is, insertion or deletion mutations) introduced in one or both lineages in the time since they diverged from one another. In sequence alignments of proteins, the degree of similarity between amino acids occupying a pa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Big O Notation
Big ''O'' notation is a mathematical notation that describes the limiting behavior of a function when the argument tends towards a particular value or infinity. Big O is a member of a family of notations invented by Paul Bachmann, Edmund Landau, and others, collectively called Bachmann–Landau notation or asymptotic notation. The letter O was chosen by Bachmann to stand for '' Ordnung'', meaning the order of approximation. In computer science, big O notation is used to classify algorithms according to how their run time or space requirements grow as the input size grows. In analytic number theory, big O notation is often used to express a bound on the difference between an arithmetical function and a better understood approximation; a famous example of such a difference is the remainder term in the prime number theorem. Big O notation is also used in many other fields to provide similar estimates. Big O notation characterizes functions according to their growth rates: d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homology (biology)
In biology, homology is similarity due to shared ancestry between a pair of structures or genes in different taxa. A common example of homologous structures is the forelimbs of vertebrates, where the wings of bats and birds, the arms of primates, the front flippers of whales and the forelegs of four-legged vertebrates like dogs and crocodiles are all derived from the same ancestral tetrapod structure. Evolutionary biology explains homologous structures adapted to different purposes as the result of descent with modification from a common ancestor. The term was first applied to biology in a non-evolutionary context by the anatomist Richard Owen in 1843. Homology was later explained by Charles Darwin's theory of evolution in 1859, but had been observed before this, from Aristotle onwards, and it was explicitly analysed by Pierre Belon in 1555. In developmental biology, organs that developed in the embryo in the same manner and from similar origins, such as from matc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Expectation Value
In probability theory, the expected value (also called expectation, expectancy, mathematical expectation, mean, average, or first moment) is a generalization of the weighted average. Informally, the expected value is the arithmetic mean of a large number of independently selected outcomes of a random variable. The expected value of a random variable with a finite number of outcomes is a weighted average of all possible outcomes. In the case of a continuum of possible outcomes, the expectation is defined by integration. In the axiomatic foundation for probability provided by measure theory, the expectation is given by Lebesgue integration. The expected value of a random variable is often denoted by , , or , with also often stylized as or \mathbb. History The idea of the expected value originated in the middle of the 17th century from the study of the so-called problem of points, which seeks to divide the stakes ''in a fair way'' between two players, who have to end t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gap Penalty
A Gap penalty is a method of scoring alignments of two or more sequences. When aligning sequences, introducing gaps in the sequences can allow an alignment algorithm to match more terms than a gap-less alignment can. However, minimizing gaps in an alignment is important to create a useful alignment. Too many gaps can cause an alignment to become meaningless. Gap penalties are used to adjust alignment scores based on the number and length of gaps. The five main types of gap penalties are constant, linear, affine, convex, and profile-based. Applications * Genetic sequence alignment - In bioinformatics, gaps are used to account for genetic mutations occurring from insertions or deletions in the sequence, sometimes referred to as ''indels''. Insertions or deletions can occur due to single mutations, unbalanced crossover in meiosis, slipped strand mispairing, and chromosomal translocation. The notion of a gap in an alignment is important in many biological applications, since the i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Motif
In biology, a sequence motif is a nucleotide or amino-acid sequence pattern that is widespread and usually assumed to be related to biological function of the macromolecule. For example, an ''N''-glycosylation site motif can be defined as ''Asn, followed by anything but Pro, followed by either Ser or Thr, followed by anything but Pro residue''. Overview When a sequence motif appears in the exon of a gene, it may encode the "structural motif" of a protein; that is a stereotypical element of the overall structure of the protein. Nevertheless, motifs need not be associated with a distinctive secondary structure. " Noncoding" sequences are not translated into proteins, and nucleic acids with such motifs need not deviate from the typical shape (e.g. the "B-form" DNA double helix). Outside of gene exons, there exist regulatory sequence motifs and motifs within the " junk", such as satellite DNA. Some of these are believed to affect the shape of nucleic acids (see for exampl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Conserved Sequence
In evolutionary biology, conserved sequences are identical or similar sequences in nucleic acids ( DNA and RNA) or proteins across species ( orthologous sequences), or within a genome ( paralogous sequences), or between donor and receptor taxa ( xenologous sequences). Conservation indicates that a sequence has been maintained by natural selection. A highly conserved sequence is one that has remained relatively unchanged far back up the phylogenetic tree, and hence far back in geological time. Examples of highly conserved sequences include the RNA components of ribosomes present in all domain (biology), domains of life, the homeobox sequences widespread amongst Eukaryotes, and the tmRNA in Bacteria. The study of sequence conservation overlaps with the fields of genomics, proteomics, evolutionary biology, phylogenetics, bioinformatics and mathematics. History The discovery of the role of DNA#History, DNA in heredity, and observations by Frederick Sanger of variation betwee ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome Project
Genome projects are scientific endeavours that ultimately aim to determine the complete genome sequence of an organism (be it an animal, a plant, a fungus, a bacterium, an archaean, a protist or a virus) and to annotate protein-coding genes and other important genome-encoded features. The genome sequence of an organism includes the collective DNA sequences of each chromosome in the organism. For a bacterium containing a single chromosome, a genome project will aim to map the sequence of that chromosome. For the human species, whose genome includes 22 pairs of autosomes and 2 sex chromosomes, a complete genome sequence will involve 46 separate chromosome sequences. The Human Genome Project is a well known example of a genome project. Genome assembly Genome assembly refers to the process of taking a large number of short DNA sequences and reassembling them to create a representation of the original chromosomes from which the DNA originated. In a shotgun sequencing project, all ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Advances In Mathematics
''Advances in Mathematics'' is a peer-reviewed scientific journal covering research on pure mathematics. It was established in 1961 by Gian-Carlo Rota. The journal publishes 18 issues each year, in three volumes. At the origin, the journal aimed at publishing articles addressed to a broader "mathematical community", and not only to mathematicians in the author's field. Herbert Busemann writes, in the preface of the first issue, "The need for expository articles addressing either all mathematicians or only those in somewhat related fields has long been felt, but little has been done outside of the USSR. The serial publication ''Advances in Mathematics'' was created in response to this demand." Abstracting and indexing The journal is abstracted and indexed in: * [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Journal Of Applied Mathematics ...
This is a list of academic journals published by Hindawi. A B C D E G H I J L M N O P Q R S T U V W References {{Reflist External linksList of journals published by Hindawi * Hindawi Hindawi or Hindavi can refer to: *Hindawi affair *Hindustani language (Hindi-Urdu), Central Indo-Aryan languages, or any language of the Hindi Belt *Something of, from, or related to Hind or Hindustan (another name of India) *Hindawi Programming Sy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |