|
Genome Project
Genome projects are scientific endeavours that ultimately aim to determine the complete genome sequence of an organism (be it an animal, a plant, a fungus, a bacterium, an archaean, a protist or a virus) and to annotate protein-coding genes and other important genome-encoded features. The genome sequence of an organism includes the collective DNA sequences of each chromosome in the organism. For a bacterium containing a single chromosome, a genome project will aim to map the sequence of that chromosome. For the human species, whose genome includes 22 pairs of autosomes and 2 sex chromosomes, a complete genome sequence will involve 46 separate chromosome sequences. The Human Genome Project is a well known example of a genome project. Genome assembly Genome assembly refers to the process of taking a large number of short DNA sequences and reassembling them to create a representation of the original chromosomes from which the DNA originated. In a shotgun sequencing project, all th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetics
Genetics is the study of genes, genetic variation, and heredity in organisms.Hartl D, Jones E (2005) It is an important branch in biology because heredity is vital to organisms' evolution. Gregor Mendel, a Moravian Augustinians, Augustinian friar working in the 19th century in Brno, was the first to study genetics scientifically. Mendel studied "trait inheritance", patterns in the way traits are handed down from parents to offspring over time. He observed that organisms (pea plants) inherit traits by way of discrete "units of inheritance". This term, still used today, is a somewhat ambiguous definition of what is referred to as a gene. Phenotypic trait, Trait inheritance and Molecular genetics, molecular inheritance mechanisms of genes are still primary principles of genetics in the 21st century, but modern genetics has expanded to study the function and behavior of genes. Gene structure and function, variation, and distribution are studied within the context of the Cell (bi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Human Genome Project
The Human Genome Project (HGP) was an international scientific research project with the goal of determining the base pairs that make up human DNA, and of identifying, mapping and sequencing all of the genes of the human genome from both a physical and a functional standpoint. It started in 1990 and was completed in 2003. It was the world's largest collaborative biological project. Planning for the project began in 1984 by the US government, and it officially launched in 1990. It was declared complete on 14 April 2003, and included about 92% of the genome. Level "complete genome" was achieved in May 2021, with only 0.3% of the bases covered by potential issues. The final gapless assembly was finished in January 2022. Funding came from the US government through the National Institutes of Health (NIH) as well as numerous other groups from around the world. A parallel project was conducted outside the government by the Celera Corporation, or Celera Genomics, which was formall ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Sequence
A nucleic acid sequence is a succession of bases within the nucleotides forming alleles within a DNA (using GACT) or RNA (GACU) molecule. This succession is denoted by a series of a set of five different letters that indicate the order of the nucleotides. By convention, sequences are usually presented from the 5' end to the 3' end. For DNA, with its double helix, there are two possible directions for the notated sequence; of these two, the sense strand is used. Because nucleic acids are normally linear (unbranched) polymers, specifying the sequence is equivalent to defining the covalent structure of the entire molecule. For this reason, the nucleic acid sequence is also termed the primary structure. The sequence represents genetic information. Biological deoxyribonucleic acid represents the information which directs the functions of an organism. Nucleic acids also have a secondary structure and tertiary structure. Primary structure is sometimes mistakenly referred to as "prim ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Annotation
In molecular biology and genetics, DNA annotation or genome annotation is the process of describing the structure and function of the components of a genome, by analyzing and interpreting them in order to extract their biological significance and understand the biological processes in which they participate. Among other things, it identifies the locations of genes and all the coding regions in a genome and determines what those genes do. Annotation is performed after a genome is sequenced and assembled, and is a necessary step in genome analysis before the sequence is deposited in a database and described in a published article. Although describing individual genes and their products or functions is sufficient to consider this description as an annotation, the depth of analysis reported in literature for different genomes vary widely, with some reports including additional information that goes beyond a simple annotation. Furthermore, due to the size and complexity of sequenced ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bioinformatics
Bioinformatics () is an interdisciplinary field of science that develops methods and Bioinformatics software, software tools for understanding biological data, especially when the data sets are large and complex. Bioinformatics uses biology, chemistry, physics, computer science, data science, computer programming, information engineering, mathematics and statistics to analyze and interpret biological data. The process of analyzing and interpreting data can sometimes be referred to as computational biology, however this distinction between the two terms is often disputed. To some, the term ''computational biology'' refers to building and using models of biological systems. Computational, statistical, and computer programming techniques have been used for In silico, computer simulation analyses of biological queries. They include reused specific analysis "pipelines", particularly in the field of genomics, such as by the identification of genes and single nucleotide polymorphis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Biology
Molecular biology is a branch of biology that seeks to understand the molecule, molecular basis of biological activity in and between Cell (biology), cells, including biomolecule, biomolecular synthesis, modification, mechanisms, and interactions. Though cells and other microscopic structures had been observed in living organisms as early as the 18th century, a detailed understanding of the mechanisms and interactions governing their behavior did not emerge until the 20th century, when technologies used in physics and chemistry had advanced sufficiently to permit their application in the biological sciences. The term 'molecular biology' was first used in 1945 by the English physicist William Astbury, who described it as an approach focused on discerning the underpinnings of biological phenomena—i.e. uncovering the physical and chemical structures and properties of biological molecules, as well as their interactions with other molecules and how these interactions explain observ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-nucleotide Polymorphism
In genetics and bioinformatics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population (e.g. 1% or more), many publications do not apply such a frequency threshold. For example, a Guanine, G nucleotide present at a specific location in a reference genome may be replaced by an Adenine, A in a minority of individuals. The two possible nucleotide variations of this SNP – G or A – are called alleles. SNPs can help explain differences in susceptibility to a wide range of diseases across a population. For example, a common SNP in the Factor H, CFH gene is associated with increased risk of age-related macular degeneration. Differences in the severity of an illness or response to treatments may also be manifestations of genetic variations caused by SNPs. For example, two ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beijing Genomics Institute
BGI Group, formerly Beijing Genomics Institute, is a Chinese genomics company with headquarters in Yantian, Shenzhen, Yantian, Shenzhen. The company was originally formed in 1999 as a genetics research center to participate in the Human Genome Project. It also sequences the genomes of other animals, plants and microorganisms. BGI has transformed from a small research institute, notable for decoding the DNA of pandas and rice plants, into a diversified company active in Cloning, animal cloning, health testing, and contract research. BGI's earlier research was continued by the Beijing Institute of Genomics, Chinese Academy of Sciences. BGI Research, the group's nonprofit division, works with the Institute of Genomics and operates the China National GeneBank under a contract with the Chinese government. BGI Genomics, a subsidiary, was listed on the Shenzhen Stock Exchange in 2017. The company is supported by several China Government Guidance Funds and State-owned enterprises of China, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Assembly
In bioinformatics, sequence assembly refers to aligning and merging fragments from a longer DNA sequence in order to reconstruct the original sequence. This is needed as DNA sequencing technology might not be able to 'read' whole genomes in one go, but rather reads small pieces of between 20 and 30,000 bases, depending on the technology used. Typically, the short fragments (reads) result from shotgun sequencing genomic DNA, or gene transcript ( ESTs). The problem of sequence assembly can be compared to taking many copies of a book, passing each of them through a shredder with a different cutter, and piecing the text of the book back together just by looking at the shredded pieces. Besides the obvious difficulty of this task, there are some extra practical issues: the original may have many repeated paragraphs, and some shreds may be modified during shredding to have typos. Excerpts from another book may also be added in, and some shreds may be completely unrecognizable. Types ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Mapping
Gene mapping or genome mapping describes the methods used to identify the location of a gene on a chromosome and the distances between genes. Gene mapping can also describe the distances between different sites within a gene. The essence of all genome mapping is to place a collection of molecular markers onto their respective positions on the genome. Molecular markers come in all forms. Genes can be viewed as one special type of genetic markers in the construction of genome maps, and mapped the same way as any other markers. In some areas of study, gene mapping contributes to the creation of new recombinants within an organism. Gene maps help describe the spatial arrangement of genes on a chromosome. Genes are designated to a specific location on a chromosome known as the locus and can be used as molecular markers to find the distance between other genes on a chromosome. Maps provide researchers with the opportunity to predict the inheritance patterns of specific traits, which ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
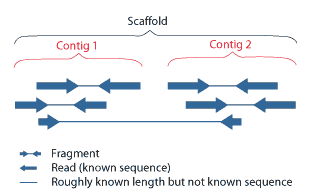
Contig
A contig (from ''contiguous'') is a set of overlapping DNA segments that together represent a consensus region of DNA.Gregory, S. ''Contig Assembly''. Encyclopedia of Life Sciences, 2005. In bottom-up sequencing projects, a contig refers to overlapping sequence data ( reads); in top-down sequencing projects, contig refers to the overlapping clones that form a physical map of the genome that is used to guide sequencing and assembly.Dear, P. H. ''Genome Mapping''. Encyclopedia of Life Sciences, 2005. . Contigs can thus refer both to overlapping DNA sequences and to overlapping physical segments (fragments) contained in clones depending on the context. Original definition of contig In 1980, Staden wrote: ''In order to make it easier to talk about our data gained by the shotgun method of sequencing we have invented the word "contig". A contig is a set of gel readings that are related to one another by overlap of their sequences. All gel readings belong to one and only one cont ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Repeated Sequence (DNA)
Repeated sequences (also known as repetitive elements, repeating units or repeats) are short or long patterns that occur in multiple copies throughout the genome. In many organisms, a significant fraction of the genomic DNA is repetitive, with over two-thirds of the sequence consisting of repetitive elements in humans. Some of these repeated sequences are necessary for maintaining important genome structures such as telomeres or centromeres. Repeated sequences are categorized into different classes depending on features such as structure, length, location, origin, and mode of multiplication. The disposition of repetitive elements throughout the genome can consist either in directly adjacent arrays called tandem repeats or in repeats dispersed throughout the genome called interspersed repeats. Tandem repeats and interspersed repeats are further categorized into subclasses based on the length of the repeated sequence and/or the mode of multiplication. While some repeated DNA seque ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |