Smith–Waterman Algorithm on:
[Wikipedia]
[Google]
[Amazon]
The Smith–Waterman algorithm performs local
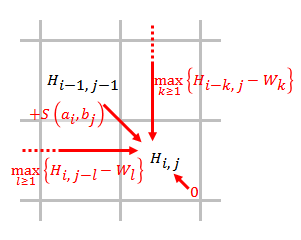 Let and be the sequences to be aligned, where and are the lengths of and respectively.
# Determine the substitution matrix and the gap penalty scheme.
#* - Similarity score of the elements that constituted the two sequences
#* - The penalty of a gap that has length
# Construct a scoring matrix and initialize its first row and first column. The size of the scoring matrix is . The matrix uses 0-based indexing.
#:
# Fill the scoring matrix using the equation below.
#:
#: where
#: is the score of aligning and ,
#: is the score if is at the end of a gap of length ,
#: is the score if is at the end of a gap of length ,
#: means there is no similarity up to and .
# Traceback. Starting at the highest score in the scoring matrix and ending at a matrix cell that has a score of 0, traceback based on the source of each score recursively to generate the best local alignment.
Let and be the sequences to be aligned, where and are the lengths of and respectively.
# Determine the substitution matrix and the gap penalty scheme.
#* - Similarity score of the elements that constituted the two sequences
#* - The penalty of a gap that has length
# Construct a scoring matrix and initialize its first row and first column. The size of the scoring matrix is . The matrix uses 0-based indexing.
#:
# Fill the scoring matrix using the equation below.
#:
#: where
#: is the score of aligning and ,
#: is the score if is at the end of a gap of length ,
#: is the score if is at the end of a gap of length ,
#: means there is no similarity up to and .
# Traceback. Starting at the highest score in the scoring matrix and ending at a matrix cell that has a score of 0, traceback based on the source of each score recursively to generate the best local alignment.
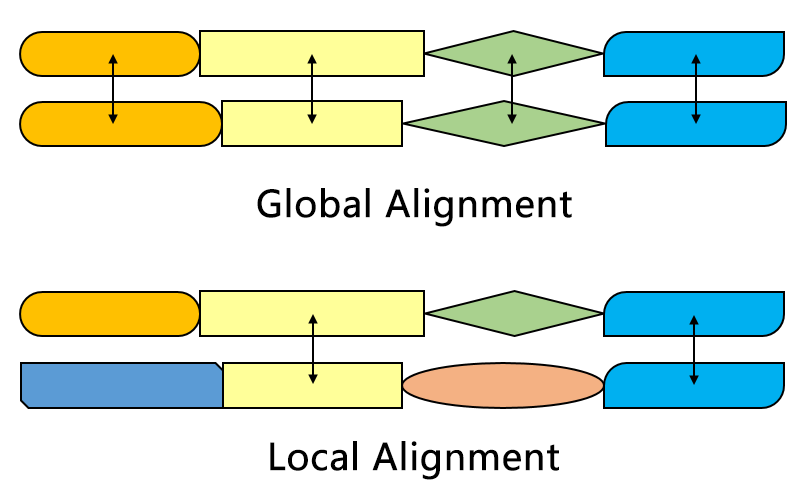 The Smith–Waterman algorithm finds the segments in two sequences that have similarities while the Needleman–Wunsch algorithm aligns two complete sequences. Therefore, they serve different purposes. Both algorithms use the concepts of a substitution matrix, a gap penalty function, a scoring matrix, and a traceback process. Three main differences are:
One of the most important distinctions is that no negative score is assigned in the scoring system of the Smith–Waterman algorithm, which enables local alignment. When any element has a score lower than zero, it means that the sequences up to this position have no similarities; this element will then be set to zero to eliminate influence from previous alignment. In this way, calculation can continue to find alignment in any position afterwards.
The initial scoring matrix of Smith–Waterman algorithm enables the alignment of any segment of one sequence to an arbitrary position in the other sequence. In Needleman–Wunsch algorithm, however, end gap penalty also needs to be considered in order to align the full sequences.
The Smith–Waterman algorithm finds the segments in two sequences that have similarities while the Needleman–Wunsch algorithm aligns two complete sequences. Therefore, they serve different purposes. Both algorithms use the concepts of a substitution matrix, a gap penalty function, a scoring matrix, and a traceback process. Three main differences are:
One of the most important distinctions is that no negative score is assigned in the scoring system of the Smith–Waterman algorithm, which enables local alignment. When any element has a score lower than zero, it means that the sequences up to this position have no similarities; this element will then be set to zero to eliminate influence from previous alignment. In this way, calculation can continue to find alignment in any position afterwards.
The initial scoring matrix of Smith–Waterman algorithm enables the alignment of any segment of one sequence to an arbitrary position in the other sequence. In Needleman–Wunsch algorithm, however, end gap penalty also needs to be considered in order to align the full sequences.
EMBOSS Water
are: gap opening = 10, gap extension = 0.5. Here we discuss two common strategies for gap penalty. See
 A linear gap penalty has the same scores for opening and extending a gap:
,
where is the cost of a single gap.
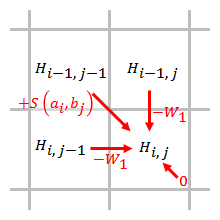
The gap penalty is directly proportional to the gap length. When linear gap penalty is used, the Smith–Waterman algorithm can be simplified to:
The simplified algorithm uses steps. When an element is being scored, only the gap penalties from the elements that are directly adjacent to this element need to be considered.
A linear gap penalty has the same scores for opening and extending a gap:
,
where is the cost of a single gap.
The gap penalty is directly proportional to the gap length. When linear gap penalty is used, the Smith–Waterman algorithm can be simplified to:
The simplified algorithm uses steps. When an element is being scored, only the gap penalties from the elements that are directly adjacent to this element need to be considered.
 The finished scoring matrix is shown below on the left. The blue color shows the highest score. An element can receive score from more than one element, each will form a different path if this element is traced back. In case of multiple highest scores, traceback should be done starting with each highest score. The traceback process is shown below on the right. The best local alignment is generated in the reverse direction.
The alignment result is:
The finished scoring matrix is shown below on the left. The blue color shows the highest score. An element can receive score from more than one element, each will form a different path if this element is traced back. In case of multiple highest scores, traceback should be done starting with each highest score. The traceback process is shown below on the right. The best local alignment is generated in the reverse direction.
The alignment result is:
UVA FASTA Downloads
This implementation includes Altivec accelerated code for
White Paper - Accelerating Intensive Applications at 10×–50× Speedup to Remove Bottlenecks in Computational Workflows
. Th
TimeLogic
DeCypher and CodeQuest systems also accelerate Smith–Waterman and Framesearch using PCIe FPGA cards. A 2011 Master's thesis includes an analysis of FPGA-based Smith–Waterman acceleration. In a 2016 publicatio
OpenCL code compiled with Xilinx SDAccel accelerates genome sequencing, beats CPU/GPU performance/W by 12-21x
a very efficient implementation was presented. Using one PCIe FPGA card equipped with a Xilinx Virtex-7 2000T FPGA, the performance per Watt level was better than CPU/GPU by 12-21x.
CUDASW++
Eleven different SW implementations on CUDA have been reported, three of which report speedups of 30X. Finally, other GPU-accelerated implementations of the Smith-Waterman can be found in
Sencel Bioinformatics
has applied for a patent covering this approach. Sencel is developing the software further and provides executables for academic use free of charge. A
similarity searching programs
Danish bioinformatics company CLC bio has achieved speed-ups of close to 200 over standard software implementations with SSE2 on an Intel 2.17 GHz Core 2 Duo CPU, according to
publicly available white paper
Accelerated version of the Smith–Waterman algorithm, on
GenCore 6
package, offered b
Biocceleration
Performance benchmarks of this software package show up to 10 fold speed acceleration relative to standard software implementation on the same processor. Currently the only company in bioinformatics to offer both SSE and FPGA solutions accelerating Smith–Waterman, CLC bio has achieved speed-ups of more than 110 over standard software implementations wit
CLC Bioinformatics Cube
The fastest implementation of the algorithm on CPUs with
JAligner
— an open source Java implementation of the Smith–Waterman algorithm
B.A.B.A.
— an applet (with source) which visually explains the algorithm
FASTA/SSEARCH
— services page at the EBI
UGENE Smith–Waterman plugin
— an open source SSEARCH compatible implementation of the algorithm with graphical interface written in C++
OPAL
— an SIMD C/C++ library for massive optimal sequence alignment
diagonalsw
— an open-source C/C++ implementation with SIMD instruction sets (notably SSE4.1) under the MIT license
SSW
— an open-source C++ library providing an API to an SIMD implementation of the Smith–Waterman algorithm under the MIT license
melodic sequence alignment
— a javascript implementation for melodic sequence alignment
DRAGMAP
A C++ port of the Illumina DRAGEN FPGA implementation {{DEFAULTSORT:Smith-Waterman algorithm Bioinformatics algorithms Computational phylogenetics Sequence alignment algorithms Dynamic programming
sequence alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural biology, structural, or evolutionary relationships between ...
; that is, for determining similar regions between two strings of nucleic acid sequence
A nucleic acid sequence is a succession of Nucleobase, bases within the nucleotides forming alleles within a DNA (using GACT) or RNA (GACU) molecule. This succession is denoted by a series of a set of five different letters that indicate the orde ...
s or protein sequence
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthe ...
s. Instead of looking at the entire
Entire may refer to:
* Entire function, a function that is holomorphic on the whole complex plane
* Entire (animal), an indication that an animal is not neutered
* Entire (botany)
This glossary of botanical terms is a list of definitions o ...
sequence, the Smith–Waterman algorithm compares segments of all possible lengths and optimizes the similarity measure
In statistics and related fields, a similarity measure or similarity function or similarity metric is a real-valued function that quantifies the similarity between two objects. Although no single definition of a similarity exists, usually such mea ...
.
The algorithm was first proposed by Temple F. Smith
Temple Ferris Smith (born March 7, 1939) is an emeritus professor in biomedical engineering who helped to develop the Smith-Waterman algorithm with Michael Waterman in 1981. The Smith-Waterman algorithm serves as the basis for multi sequence co ...
and Michael S. Waterman in 1981. Like the Needleman–Wunsch algorithm
The Needleman–Wunsch algorithm is an algorithm used in bioinformatics to align protein or nucleotide sequences. It was one of the first applications of dynamic programming to compare biological sequences. The algorithm was developed by Saul ...
, of which it is a variation, Smith–Waterman is a dynamic programming algorithm. As such, it has the desirable property that it is guaranteed to find the optimal local alignment with respect to the scoring system being used (which includes the substitution matrix
In bioinformatics and evolutionary biology, a substitution matrix describes the frequency at which a character in a Nucleic acid sequence, nucleotide sequence or a Protein primary structure, protein sequence changes to other character states ove ...
and the gap-scoring scheme). The main difference to the Needleman–Wunsch algorithm
The Needleman–Wunsch algorithm is an algorithm used in bioinformatics to align protein or nucleotide sequences. It was one of the first applications of dynamic programming to compare biological sequences. The algorithm was developed by Saul ...
is that negative scoring matrix cells are set to zero. Traceback procedure starts at the highest scoring matrix cell and proceeds until a cell with score zero is encountered, yielding the highest scoring local alignment. Because of its quadratic time complexity, it often cannot be practically applied to large-scale problems and is replaced in favor of computationally more efficient alternatives such as (Gotoh, 1982), ( Altschul and Erickson, 1986), and (Myers and Miller, 1988).
History
In 1970, Saul B. Needleman and Christian D. Wunsch proposed a heuristic homology algorithm for sequence alignment, also referred to as the Needleman–Wunsch algorithm. It is a global alignment algorithm that requires calculation steps ( and are the lengths of the two sequences being aligned). It uses the iterative calculation of a matrix for the purpose of showing global alignment. In the following decade, Sankoff, Reichert, Beyer and others formulated alternative heuristic algorithms for analyzing gene sequences. Sellers introduced a system for measuring sequence distances. In 1976, Waterman et al. added the concept of gaps into the original measurement system. In 1981, Smith and Waterman published their Smith–Waterman algorithm for calculating local alignment. The Smith–Waterman algorithm is fairly demanding of time: To align two sequences of lengths and , time is required. Gotoh and Altschul optimized the algorithm to steps. The space complexity was optimized by Myers and Miller from to (linear), where is the length of the shorter sequence, for the case where only one of the many possible optimal alignments is desired. Chowdhury, Le, and Ramachandran later optimized the cache performance of the algorithm while keeping the space usage linear in the total length of the input sequences.Motivation
In recent years,genome project
Genome projects are scientific endeavours that ultimately aim to determine the complete genome sequence of an organism (be it an animal, a plant, a fungus, a bacterium, an archaean, a protist or a virus) and to annotate protein-coding genes and o ...
s conducted on a variety of organisms generated massive amounts of sequence data for genes and proteins, which requires computational analysis. Sequence alignment shows the relations between genes or between proteins, leading to a better understanding of their homology and functionality. Sequence alignment can also reveal conserved domains and motifs.
One motivation for local alignment is the difficulty of obtaining correct alignments in regions of low similarity between distantly related biological sequences, because mutations have added too much 'noise' over evolutionary time to allow for a meaningful comparison of those regions. Local alignment avoids such regions altogether and focuses on those with a positive score, i.e. those with an evolutionarily conserved signal of similarity. A prerequisite for local alignment is a negative expectation score. The expectation score is defined as the average score that the scoring system (substitution matrix
In bioinformatics and evolutionary biology, a substitution matrix describes the frequency at which a character in a Nucleic acid sequence, nucleotide sequence or a Protein primary structure, protein sequence changes to other character states ove ...
and gap penalties) would yield for a random sequence.
Another motivation for using local alignments is that there is a reliable statistical model (developed by Karlin and Altschul) for optimal local alignments. The alignment of unrelated sequences tends to produce optimal local alignment scores which follow an extreme value distribution. This property allows programs to produce an expectation value
In probability theory, the expected value (also called expectation, expectancy, expectation operator, mathematical expectation, mean, expectation value, or first moment) is a generalization of the weighted average. Informally, the expected va ...
for the optimal local alignment of two sequences, which is a measure of how often two unrelated sequences would produce an optimal local alignment whose score is greater than or equal to the observed score. Very low expectation values indicate that the two sequences in question might be homologous, meaning they might share a common ancestor.
Algorithm
 Let and be the sequences to be aligned, where and are the lengths of and respectively.
# Determine the substitution matrix and the gap penalty scheme.
#* - Similarity score of the elements that constituted the two sequences
#* - The penalty of a gap that has length
# Construct a scoring matrix and initialize its first row and first column. The size of the scoring matrix is . The matrix uses 0-based indexing.
#:
# Fill the scoring matrix using the equation below.
#:
#: where
#: is the score of aligning and ,
#: is the score if is at the end of a gap of length ,
#: is the score if is at the end of a gap of length ,
#: means there is no similarity up to and .
# Traceback. Starting at the highest score in the scoring matrix and ending at a matrix cell that has a score of 0, traceback based on the source of each score recursively to generate the best local alignment.
Let and be the sequences to be aligned, where and are the lengths of and respectively.
# Determine the substitution matrix and the gap penalty scheme.
#* - Similarity score of the elements that constituted the two sequences
#* - The penalty of a gap that has length
# Construct a scoring matrix and initialize its first row and first column. The size of the scoring matrix is . The matrix uses 0-based indexing.
#:
# Fill the scoring matrix using the equation below.
#:
#: where
#: is the score of aligning and ,
#: is the score if is at the end of a gap of length ,
#: is the score if is at the end of a gap of length ,
#: means there is no similarity up to and .
# Traceback. Starting at the highest score in the scoring matrix and ending at a matrix cell that has a score of 0, traceback based on the source of each score recursively to generate the best local alignment.
Explanation
Smith–Waterman algorithm aligns two sequences by matches/mismatches (also known as substitutions), insertions, and deletions. Both insertions and deletions are the operations that introduce gaps, which are represented by dashes. The Smith–Waterman algorithm has several steps: # Determine the substitution matrix and the gap penalty scheme. A substitution matrix assigns each pair of bases or amino acids a score for match or mismatch. Usually matches get positive scores, whereas mismatches get relatively lower scores. A gap penalty function determines the score cost for opening or extending gaps. It is suggested that users choose the appropriate scoring system based on the goals. In addition, it is also a good practice to try different combinations of substitution matrices and gap penalties. # Initialize the scoring matrix. The dimensions of the scoring matrix are 1+length of each sequence respectively. All the elements of the first row and the first column are set to 0. The extra first row and first column make it possible to align one sequence to another at any position, and setting them to 0 makes the terminal gap free from penalty. # Scoring. Score each element from left to right, top to bottom in the matrix, considering the outcomes of substitutions (diagonal scores) or adding gaps (horizontal and vertical scores). If none of the scores are positive, this element gets a 0. Otherwise the highest score is used and the source of that score is recorded. # Traceback. Starting at the element with the highest score, traceback based on the source of each score recursively, until 0 is encountered. The segments that have the highest similarity score based on the given scoring system is generated in this process. To obtain the second best local alignment, apply the traceback process starting at the second highest score outside the trace of the best alignment.Comparison with the Needleman–Wunsch algorithm
 The Smith–Waterman algorithm finds the segments in two sequences that have similarities while the Needleman–Wunsch algorithm aligns two complete sequences. Therefore, they serve different purposes. Both algorithms use the concepts of a substitution matrix, a gap penalty function, a scoring matrix, and a traceback process. Three main differences are:
One of the most important distinctions is that no negative score is assigned in the scoring system of the Smith–Waterman algorithm, which enables local alignment. When any element has a score lower than zero, it means that the sequences up to this position have no similarities; this element will then be set to zero to eliminate influence from previous alignment. In this way, calculation can continue to find alignment in any position afterwards.
The initial scoring matrix of Smith–Waterman algorithm enables the alignment of any segment of one sequence to an arbitrary position in the other sequence. In Needleman–Wunsch algorithm, however, end gap penalty also needs to be considered in order to align the full sequences.
The Smith–Waterman algorithm finds the segments in two sequences that have similarities while the Needleman–Wunsch algorithm aligns two complete sequences. Therefore, they serve different purposes. Both algorithms use the concepts of a substitution matrix, a gap penalty function, a scoring matrix, and a traceback process. Three main differences are:
One of the most important distinctions is that no negative score is assigned in the scoring system of the Smith–Waterman algorithm, which enables local alignment. When any element has a score lower than zero, it means that the sequences up to this position have no similarities; this element will then be set to zero to eliminate influence from previous alignment. In this way, calculation can continue to find alignment in any position afterwards.
The initial scoring matrix of Smith–Waterman algorithm enables the alignment of any segment of one sequence to an arbitrary position in the other sequence. In Needleman–Wunsch algorithm, however, end gap penalty also needs to be considered in order to align the full sequences.
Substitution matrix
Each base substitution or amino acid substitution is assigned a score. In general, matches are assigned positive scores, and mismatches are assigned relatively lower scores. Take DNA sequence as an example. If matches get +1, mismatches get -1, then the substitution matrix is: This substitution matrix can be described as: Different base substitutions or amino acid substitutions can have different scores. The substitution matrix of amino acids is usually more complicated than that of the bases. See PAM,BLOSUM
In bioinformatics, the BLOSUM (BLOcks SUbstitution Matrix) matrix is a substitution matrix used for sequence alignment of proteins. BLOSUM matrices are used to score alignments between evolutionarily divergent protein sequences. They are based o ...
.
Gap penalty
Gap penalty designates scores for insertion or deletion. A simple gap penalty strategy is to use fixed score for each gap. In biology, however, the score needs to be counted differently for practical reasons. On one hand, partial similarity between two sequences is a common phenomenon; on the other hand, a single gene mutation event can result in insertion of a single long gap. Therefore, connected gaps forming a long gap usually is more favored than multiple scattered, short gaps. In order to take this difference into consideration, the concepts of gap opening and gap extension have been added to the scoring system. The gap opening score is usually higher than the gap extension score. For instance, the default parameters iEMBOSS Water
are: gap opening = 10, gap extension = 0.5. Here we discuss two common strategies for gap penalty. See
Gap penalty A Gap penalty is a method of scoring alignments of two or more sequences. When aligning sequences, introducing gaps in the sequences can allow an alignment algorithm to match more terms than a gap-less alignment can. However, minimizing gaps in an a ...
for more strategies.
Let be the gap penalty function for a gap of length :
Linear
 A linear gap penalty has the same scores for opening and extending a gap:
,
where is the cost of a single gap.
The gap penalty is directly proportional to the gap length. When linear gap penalty is used, the Smith–Waterman algorithm can be simplified to:
The simplified algorithm uses steps. When an element is being scored, only the gap penalties from the elements that are directly adjacent to this element need to be considered.
A linear gap penalty has the same scores for opening and extending a gap:
,
where is the cost of a single gap.
The gap penalty is directly proportional to the gap length. When linear gap penalty is used, the Smith–Waterman algorithm can be simplified to:
The simplified algorithm uses steps. When an element is being scored, only the gap penalties from the elements that are directly adjacent to this element need to be considered.
Affine
An affine gap penalty considers gap opening and extension separately: , where is the gap opening penalty, and is the gap extension penalty. For example, the penalty for a gap of length 2 is . An arbitrary gap penalty was used in the original Smith–Waterman algorithm paper. It uses steps, therefore is quite demanding of time. Gotoh optimized the steps for an affine gap penalty to , but the optimized algorithm only attempts to find one optimal alignment, and the optimal alignment is not guaranteed to be found. Altschul modified Gotoh's algorithm to find all optimal alignments while maintaining the computational complexity. Later, Myers and Miller pointed out that Gotoh and Altschul's algorithm can be further modified based on the method that was published by Hirschberg in 1975, and applied this method. Myers and Miller's algorithm can align two sequences using space, with being the length of the shorter sequence. Chowdhury, Le, and Ramachandran later showed how to run Gotoh's algorithm cache-efficiently in linear space using a different recursive divide-and-conquer strategy than the one used by Hirschberg. The resulting algorithm runs faster than Myers and Miller's algorithm in practice due to its superior cache performance.Gap penalty example
Take the alignment of sequences and as an example. When linear gap penalty function is used, the result is (Alignments performed by EMBOSS Water. Substitution matrix is DNAfull (similarity score: +5 for matching characters otherwise -4). Gap opening and extension are 0.0 and 1.0 respectively): When affine gap penalty is used, the result is (Gap opening and extension are 5.0 and 1.0 respectively): This example shows that an affine gap penalty can help avoid scattered small gaps.Scoring matrix
The function of the scoring matrix is to conduct one-to-one comparisons between all components in two sequences and record the optimal alignment results. The scoring process reflects the concept of dynamic programming. The final optimal alignment is found by iteratively expanding the growing optimal alignment. In other words, the current optimal alignment is generated by deciding which path (match/mismatch or inserting gap) gives the highest score from the previous optimal alignment. The size of the matrix is the length of one sequence plus 1 by the length of the other sequence plus 1. The additional first row and first column serve the purpose of aligning one sequence to any positions in the other sequence. Both the first line and the first column are set to 0 so that end gap is not penalized. The initial scoring matrix is:Example
Take the alignment of DNA sequences and as an example. Use the following scheme: * Substitution matrix: * Gap penalty: (a linear gap penalty of ) Initialize and fill the scoring matrix, shown as below. This figure shows the scoring process of the first three elements. The yellow color indicates the bases that are being considered. The red color indicates the highest possible score for the cell being scored. The finished scoring matrix is shown below on the left. The blue color shows the highest score. An element can receive score from more than one element, each will form a different path if this element is traced back. In case of multiple highest scores, traceback should be done starting with each highest score. The traceback process is shown below on the right. The best local alignment is generated in the reverse direction.
The alignment result is:
The finished scoring matrix is shown below on the left. The blue color shows the highest score. An element can receive score from more than one element, each will form a different path if this element is traced back. In case of multiple highest scores, traceback should be done starting with each highest score. The traceback process is shown below on the right. The best local alignment is generated in the reverse direction.
The alignment result is:
Implementation
An implementation of the Smith–Waterman Algorithm, SSEARCH, is available in theFASTA
FASTA is a DNA and protein sequence alignment software package first described by David J. Lipman and William R. Pearson in 1985. Its legacy is the FASTA format which is now ubiquitous in bioinformatics.
History
The original FASTA program ...
sequence analysis package froUVA FASTA Downloads
This implementation includes Altivec accelerated code for
PowerPC
PowerPC (with the backronym Performance Optimization With Enhanced RISC – Performance Computing, sometimes abbreviated as PPC) is a reduced instruction set computer (RISC) instruction set architecture (ISA) created by the 1991 Apple Inc., App ...
G4 and G5 processors that speeds up comparisons 10–20-fold, using a modification of the Wozniak, 1997 approach, and an SSE2 vectorization developed by Farrar making optimal protein sequence database
In the field of bioinformatics, a sequence database is a type of biological database that is composed of a large collection of computerized ("Digital data, digital") nucleic acid sequences, protein sequences, or other polymer sequences stored on a ...
searches quite practical. A library, SSW, extends Farrar's implementation to return alignment information in addition to the optimal Smith–Waterman score.
Accelerated versions
FPGA
Cray
Cray Inc., a subsidiary of Hewlett Packard Enterprise, is an American supercomputer manufacturer headquartered in Seattle, Washington. It also manufactures systems for data storage and analytics. Several Cray supercomputer systems are listed ...
demonstrated acceleration of the Smith–Waterman algorithm using a reconfigurable computing
Reconfigurable computing is a computer architecture combining some of the flexibility of software with the high performance of hardware by processing with flexible hardware platforms like FPGA, field-programmable gate arrays (FPGAs). The princip ...
platform based on FPGA
A field-programmable gate array (FPGA) is a type of configurable integrated circuit that can be repeatedly programmed after manufacturing. FPGAs are a subset of logic devices referred to as programmable logic devices (PLDs). They consist of a ...
chips, with results showing up to 28x speed-up over standard microprocessor-based solutions. Another FPGA-based version of the Smith–Waterman algorithm shows FPGA (Virtex-4) speedups up to 100xFPGA 100x Papers: , , and over a 2.2 GHz Opteron processor.Progeniq Pte. Ltd.,White Paper - Accelerating Intensive Applications at 10×–50× Speedup to Remove Bottlenecks in Computational Workflows
. Th
TimeLogic
DeCypher and CodeQuest systems also accelerate Smith–Waterman and Framesearch using PCIe FPGA cards. A 2011 Master's thesis includes an analysis of FPGA-based Smith–Waterman acceleration. In a 2016 publicatio
OpenCL code compiled with Xilinx SDAccel accelerates genome sequencing, beats CPU/GPU performance/W by 12-21x
a very efficient implementation was presented. Using one PCIe FPGA card equipped with a Xilinx Virtex-7 2000T FPGA, the performance per Watt level was better than CPU/GPU by 12-21x.
GPU
Lawrence Livermore National Laboratory
Lawrence Livermore National Laboratory (LLNL) is a Federally funded research and development centers, federally funded research and development center in Livermore, California, United States. Originally established in 1952, the laboratory now i ...
and the United States (US) Department of Energy's Joint Genome Institute
The Joint Genome Institute (JGI) is a scientific user facility for integrative genomic science at Lawrence Berkeley National Laboratory. The mission of the JGI is to advance genomics research in support of the United States Department of Energy ...
implemented an accelerated version of Smith–Waterman local sequence alignment searches using graphics processing units
A graphics processing unit (GPU) is a specialized electronic circuit designed for digital image processing and to accelerate computer graphics, being present either as a discrete video card or embedded on motherboards, mobile phones, personal co ...
(GPUs) with preliminary results showing a 2x speed-up over software implementations. A similar method has already been implemented in the Biofacet software since 1997, with the same speed-up factor.
Several GPU
A graphics processing unit (GPU) is a specialized electronic circuit designed for digital image processing and to accelerate computer graphics, being present either as a discrete video card or embedded on motherboards, mobile phones, personal ...
implementations of the algorithm in NVIDIA
Nvidia Corporation ( ) is an American multinational corporation and technology company headquartered in Santa Clara, California, and incorporated in Delaware. Founded in 1993 by Jensen Huang (president and CEO), Chris Malachowsky, and Curti ...
's CUDA
In computing, CUDA (Compute Unified Device Architecture) is a proprietary parallel computing platform and application programming interface (API) that allows software to use certain types of graphics processing units (GPUs) for accelerated gene ...
C platform are also available. When compared to the best known CPU implementation (using SIMD instructions on the x86 architecture), by Farrar, the performance tests of this solution using a single NVidia GeForce 8800 GTX card show a slight increase in performance for smaller sequences, but a slight decrease in performance for larger ones. However, the same tests running on dual NVidia GeForce 8800 GTX cards are almost twice as fast as the Farrar implementation for all sequence sizes tested.
A newer GPU CUDA implementation of SW is now available that is faster than previous versions and also removes limitations on query lengths. SeCUDASW++
Eleven different SW implementations on CUDA have been reported, three of which report speedups of 30X. Finally, other GPU-accelerated implementations of the Smith-Waterman can be found in
NVIDIA Parabricks
Parabricks company started at the University of Michigan by Mehrzad Samadi, Ankit Sethia, and Scott Mahlke. It was acquired by Nvidia in 2020.
Nvidia Parabricks is a suite of free software for genome analysis developed by Nvidia, designed to ...
, NVIDIA's software suite for genome analysis.
SIMD
In 2000, a fast implementation of the Smith–Waterman algorithm using the ''single instruction, multiple data'' (SIMD
Single instruction, multiple data (SIMD) is a type of parallel computer, parallel processing in Flynn's taxonomy. SIMD describes computers with multiple processing elements that perform the same operation on multiple data points simultaneousl ...
) technology available in Intel
Intel Corporation is an American multinational corporation and technology company headquartered in Santa Clara, California, and Delaware General Corporation Law, incorporated in Delaware. Intel designs, manufactures, and sells computer compo ...
Pentium
Pentium is a series of x86 architecture-compatible microprocessors produced by Intel from 1993 to 2023. The Pentium (original), original Pentium was Intel's fifth generation processor, succeeding the i486; Pentium was Intel's flagship proce ...
MMX processors and similar technology was described in a publication by Rognes and Seeberg. In contrast to the Wozniak (1997) approach, the new implementation was based on vectors parallel with the query sequence, not diagonal vectors. The companSencel Bioinformatics
has applied for a patent covering this approach. Sencel is developing the software further and provides executables for academic use free of charge. A
SSE2
SSE2 (Streaming SIMD Extensions 2) is one of the Intel SIMD (Single Instruction, Multiple Data) processor supplementary instruction sets introduced by Intel with the initial version of the Pentium 4 in 2000. SSE2 instructions allow the use of ...
vectorization of the algorithm (Farrar, 2007) is now available providing an 8-16-fold speedup on Intel/AMD processors with SSE2 extensions. When running on Intel processor using the Core microarchitecture the SSE2 implementation achieves a 20-fold increase. Farrar's SSE2 implementation is available as the SSEARCH program in the FASTA
FASTA is a DNA and protein sequence alignment software package first described by David J. Lipman and William R. Pearson in 1985. Its legacy is the FASTA format which is now ubiquitous in bioinformatics.
History
The original FASTA program ...
sequence comparison package. The SSEARCH is included in the European Bioinformatics Institute
The European Bioinformatics Institute (EMBL-EBI) is an intergovernmental organization (IGO) which, as part of the European Molecular Biology Laboratory (EMBL) family, focuses on research and services in bioinformatics. It is located on the Wel ...
's suite osimilarity searching programs
Danish bioinformatics company CLC bio has achieved speed-ups of close to 200 over standard software implementations with SSE2 on an Intel 2.17 GHz Core 2 Duo CPU, according to
publicly available white paper
Accelerated version of the Smith–Waterman algorithm, on
Intel
Intel Corporation is an American multinational corporation and technology company headquartered in Santa Clara, California, and Delaware General Corporation Law, incorporated in Delaware. Intel designs, manufactures, and sells computer compo ...
and Advanced Micro Devices
Advanced Micro Devices, Inc. (AMD) is an American multinational corporation and technology company headquartered in Santa Clara, California and maintains significant operations in Austin, Texas. AMD is a Information technology, hardware and F ...
(AMD) based Linux
Linux ( ) is a family of open source Unix-like operating systems based on the Linux kernel, an kernel (operating system), operating system kernel first released on September 17, 1991, by Linus Torvalds. Linux is typically package manager, pac ...
servers, is supported by thGenCore 6
package, offered b
Biocceleration
Performance benchmarks of this software package show up to 10 fold speed acceleration relative to standard software implementation on the same processor. Currently the only company in bioinformatics to offer both SSE and FPGA solutions accelerating Smith–Waterman, CLC bio has achieved speed-ups of more than 110 over standard software implementations wit
CLC Bioinformatics Cube
The fastest implementation of the algorithm on CPUs with
SSSE3
Supplemental Streaming SIMD Extensions 3 (SSSE3 or SSE3S) is a SIMD instruction set created by Intel and is the fourth iteration of the SSE technology.
History
SSSE3 was first introduced with Intel processors based on the Core microarchitect ...
can be found the SWIPE software (Rognes, 2011), which is available under the GNU Affero General Public License
The GNU Affero General Public License (GNU AGPL) is a free, copyleft license published by the Free Software Foundation in November 2007, and based on the GNU GPL version 3 and the ''Affero General Public License'' (non-GNU).
It is intended fo ...
. In parallel, this software compares residues from sixteen different database sequences to one query residue. Using a 375 residue query sequence a speed of 106 billion cell updates per second (GCUPS) was achieved on a dual Intel Xeon
Xeon (; ) is a brand of x86 microprocessors designed, manufactured, and marketed by Intel, targeted at the non-consumer workstation, server, and embedded markets. It was introduced in June 1998. Xeon processors are based on the same archite ...
X5650 six-core processor system, which is over six times more rapid than software based on Farrar's 'striped' approach. It is faster than BLAST when using the BLOSUM50 matrix.
An implementation of Smith–Waterman named ''diagonalsw'', in C and C++, uses SIMD instruction sets ( SSE4.1 for the x86 platform and AltiVec for the PowerPC platform). It is released under an open-source MIT License
The MIT License is a permissive software license originating at the Massachusetts Institute of Technology (MIT) in the late 1980s. As a permissive license, it puts very few restrictions on reuse and therefore has high license compatibility.
Unl ...
.
Cell Broadband Engine
In 2008, Farrar described a port of the Striped Smith–Waterman to theCell Broadband Engine
The Cell Broadband Engine (Cell/B.E.) is a 64-bit multi-core processor and microarchitecture developed by Sony, Toshiba, and IBM—an alliance known as "STI". It combines a general-purpose PowerPC core, called the Power Processing Element (PPE), ...
and reported speeds of 32 and 12 GCUPS on an IBM QS20 blade and a Sony PlayStation 3
The PlayStation 3 (PS3) is a home video game console developed and marketed by Sony Computer Entertainment (SCE). It is the successor to the PlayStation 2, and both are part of the PlayStation brand of consoles. The PS3 was first released on ...
, respectively.
Limitations
Fast expansion of genetic data challenges speed of current DNA sequence alignment algorithms. Essential needs for an efficient and accurate method for DNA variant discovery demand innovative approaches for parallel processing in real time.See also
*Bioinformatics
Bioinformatics () is an interdisciplinary field of science that develops methods and Bioinformatics software, software tools for understanding biological data, especially when the data sets are large and complex. Bioinformatics uses biology, ...
*Sequence alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural biology, structural, or evolutionary relationships between ...
*Sequence mining
Sequential pattern mining is a topic of data mining concerned with finding statistically relevant patterns between data examples where the values are delivered in a sequence. It is usually presumed that the values are discrete, and thus time serie ...
*Needleman–Wunsch algorithm
The Needleman–Wunsch algorithm is an algorithm used in bioinformatics to align protein or nucleotide sequences. It was one of the first applications of dynamic programming to compare biological sequences. The algorithm was developed by Saul ...
*Levenshtein distance
In information theory, linguistics, and computer science, the Levenshtein distance is a string metric for measuring the difference between two sequences. The Levenshtein distance between two words is the minimum number of single-character edits (i ...
* BLAST
*FASTA
FASTA is a DNA and protein sequence alignment software package first described by David J. Lipman and William R. Pearson in 1985. Its legacy is the FASTA format which is now ubiquitous in bioinformatics.
History
The original FASTA program ...
References
External links
JAligner
— an open source Java implementation of the Smith–Waterman algorithm
B.A.B.A.
— an applet (with source) which visually explains the algorithm
FASTA/SSEARCH
— services page at the EBI
UGENE Smith–Waterman plugin
— an open source SSEARCH compatible implementation of the algorithm with graphical interface written in C++
OPAL
— an SIMD C/C++ library for massive optimal sequence alignment
diagonalsw
— an open-source C/C++ implementation with SIMD instruction sets (notably SSE4.1) under the MIT license
SSW
— an open-source C++ library providing an API to an SIMD implementation of the Smith–Waterman algorithm under the MIT license
melodic sequence alignment
— a javascript implementation for melodic sequence alignment
DRAGMAP
A C++ port of the Illumina DRAGEN FPGA implementation {{DEFAULTSORT:Smith-Waterman algorithm Bioinformatics algorithms Computational phylogenetics Sequence alignment algorithms Dynamic programming