|
Single-molecule Experiment
A single-molecule experiment is an experiment that investigates the properties of individual molecules. Single-molecule studies may be contrasted with measurements on an ensemble or bulk collection of molecules, where the individual behavior of molecules cannot be distinguished, and only average characteristics can be measured. Since many measurement techniques in biology, chemistry, and physics are not sensitive enough to observe single molecules, single-molecule fluorescence techniques (that have emerged since the 1990s for probing various processes on the level of individual molecules) caused a lot of excitement, since these supplied many new details on the measured processes that were not accessible in the past. Indeed, since the 1990s, many techniques for probing individual molecules have been developed. The first single-molecule experiments were patch clamp experiments performed in the 1970s, but these were limited to studying ion channels. Today, systems investigated using s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-particle Tracking
Single-particle tracking (SPT) is the observation of the motion of individual particles within a medium. The coordinates time series, which can be either in two dimensions (''x'', ''y'') or in three dimensions (''x'', ''y'', ''z''), is referred to as a ''trajectory''. The trajectory is typically analyzed using statistical methods to extract information about the underlying dynamics of the particle. These dynamics can reveal information about the type of transport being observed (e.g., thermal or active), the medium where the particle is moving, and interactions with other particles. In the case of random motion, trajectory analysis can be used to measure the diffusion coefficient. Applications In life sciences, single-particle tracking is broadly used to quantify the dynamics of molecules/proteins in live cells (of bacteria, yeast, mammalian cells and live ''Drosophila'' embryos). It has been extensively used to study the transcription factor dynamics in live cells. This method h ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Magnetic Tweezers
Magnetism is the class of physical attributes that are mediated by a magnetic field, which refers to the capacity to induce attractive and repulsive phenomena in other entities. Electric currents and the magnetic moments of elementary particles give rise to a magnetic field, which acts on other currents and magnetic moments. Magnetism is one aspect of the combined phenomena of electromagnetism. The most familiar effects occur in ferromagnetic materials, which are strongly attracted by magnetic fields and can be magnetized to become permanent magnets, producing magnetic fields themselves. Demagnetizing a magnet is also possible. Only a few substances are ferromagnetic; the most common ones are iron, cobalt, and nickel and their alloys. The rare-earth metals neodymium and samarium are less common examples. The prefix ' refers to iron because permanent magnetism was first observed in lodestone, a form of natural iron ore called magnetite, Fe3O4. All substances exhibit som ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Force Spectroscopy
Force spectroscopy is a set of techniques for the study of the interactions and the binding forces between individual molecules. These methods can be used to measure the mechanical properties of single polymer molecules or proteins, or individual chemical bonds. The name "force spectroscopy", although widely used in the scientific community, is somewhat misleading, because there is no true matter-radiation interaction. Techniques that can be used to perform force spectroscopy include atomic force microscopy, optical tweezers, magnetic tweezers, acoustic force spectroscopy, microneedles, and biomembranes. Force spectroscopy measures the behavior of a molecule under stretching or torsional mechanical force. In this way a great deal has been learned in recent years about the mechanochemical coupling in the enzymes responsible for muscle contraction, transport in the cell, energy generation (F1-ATPase), DNA replication and transcription (polymerases), DNA unknotting and unwinding ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-molecule Magnet
A single-molecule magnet (SMM) is a metal-organic compound that has superparamagnetic behavior below a certain blocking temperature at the molecular scale. In this temperature range, a SMM exhibits magnetic hysteresis of purely molecular origin.Introduction to Molecular Magnetism by Dr. Joris van Slageren. In contrast to conventional bulk s and s, collective long-range magnetic ordering of |
Reduced Dimensions Form
In biophysics and related fields, reduced dimension forms (RDFs) are unique on-off mechanisms for random walks that generate two-state trajectories (see Fig. 1 for an example of a RDF and Fig. 2 for an example of a two-state trajectory). It has been shown that RDFs solve two-state trajectories, since only one RDF can be constructed from the data, where this property does not hold for on-off kinetic schemes, where many kinetic schemes can be constructed from a particular two-state trajectory (even from an ideal on-off trajectory). Two-state time trajectories are very common in measurements in chemistry, physics, and the biophysics of individual molecules (e.g. measurements of protein dynamics and DNA and RNA dynamics, [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Kinetic Scheme
In physics, chemistry and related fields, a kinetic scheme is a network of states and connections between them representing the scheme of a dynamical process. Usually a kinetic scheme represents a Markovian process, while for non-Markovian processes generalized kinetic schemes are used. Figure 1 shows an illustration of a kinetic scheme. A Markovian kinetic scheme Mathematical description A kinetic scheme is a network (a directed graph) of distinct states (although repetition of states may occur and this depends on the system), where each pair of states ''i'' and ''j'' are associated with directional rates, A_ (and A_). It is described with a master equation: a first-order differential equation for the probability \vec of a system to occupy each one its states at time ''t'' (element ''i'' represents state ''i''). Written in a matrix form, this states: \frac=\mathbf\vec, where \mathbf is the matrix of connections (rates) A_. In a Markovian kinetic scheme the connections are c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fluorescence Resonance Energy Transfer
Fluorescence is the emission of light by a substance that has absorbed light or other electromagnetic radiation. It is a form of luminescence. In most cases, the emitted light has a longer wavelength, and therefore a lower photon energy, than the absorbed radiation. A perceptible example of fluorescence occurs when the absorbed radiation is in the ultraviolet region of the electromagnetic spectrum (invisible to the human eye), while the emitted light is in the visible region; this gives the fluorescent substance a distinct color that can only be seen when the substance has been exposed to UV light. Fluorescent materials cease to glow nearly immediately when the radiation source stops, unlike phosphorescent materials, which continue to emit light for some time after. Fluorescence has many practical applications, including mineralogy, gemology, medicine, chemical sensors (fluorescence spectroscopy), fluorescent labelling, dyes, biological detectors, cosmic-ray detection, vacuum ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
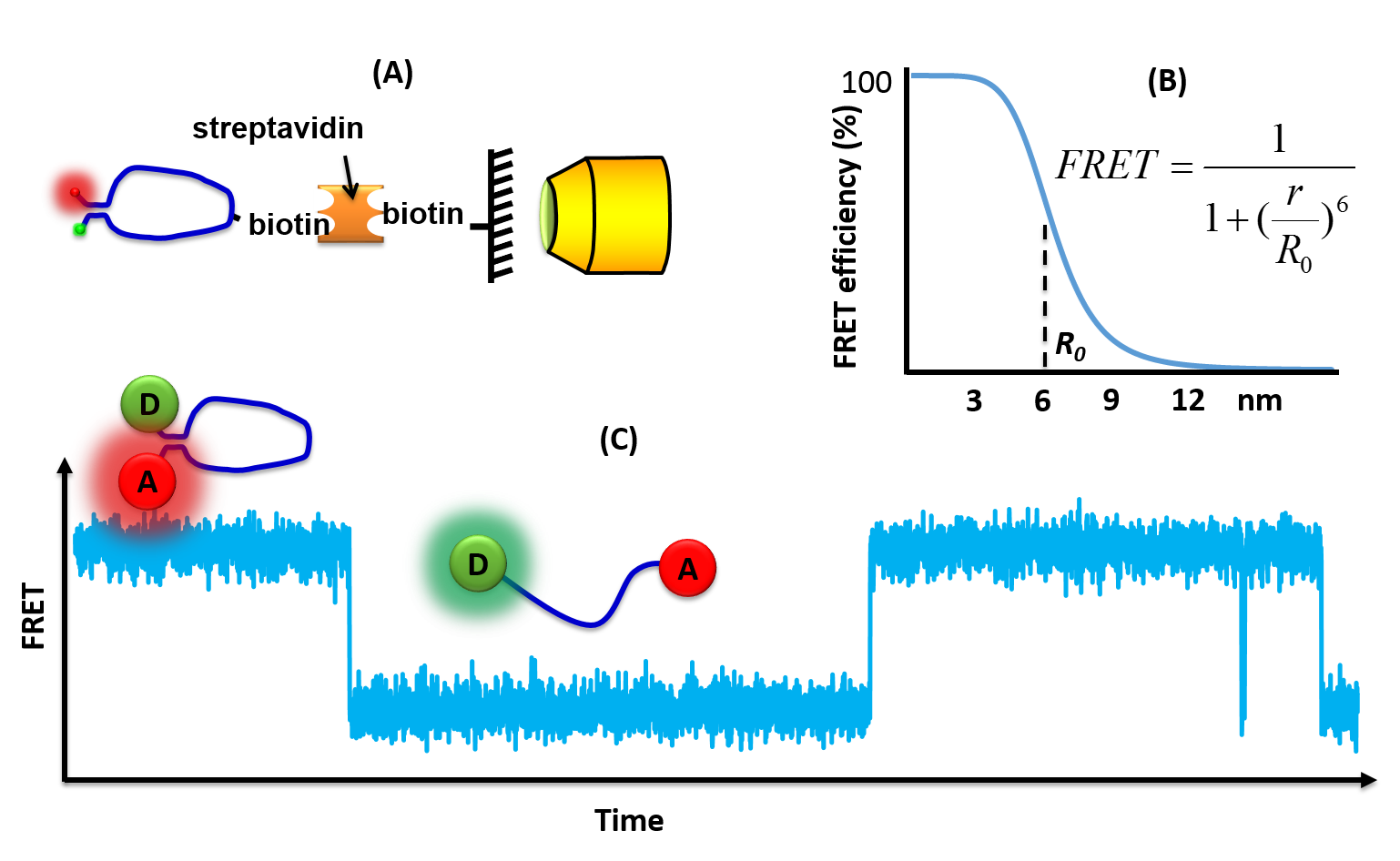
SmFRET
Single molecule fluorescence (or Förster) resonance energy transfer (or smFRET) is a biophysical technique used to measure distances at the 1-10 nanometer scale in single molecules, typically biomolecules. It is an application of FRET wherein a pair of donor and acceptor fluorophores are excited and detected on a single molecule level. In contrast to "ensemble FRET" which provides the FRET signal of a high number of molecules, single-molecule FRET is able to resolve the FRET signal of each individual molecule. The variation of the smFRET signal is useful to reveal kinetic information that an ensemble measurement cannot provide, especially when the system is under equilibrium. Heterogeneity among different molecules can also be observed. This method has been applied in many measurements of biomolecular dynamics such as DNA/RNA/protein folding/unfolding and other conformational changes, and intermolecular dynamics such as reaction, binding, adsorption, and desorption that are partic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microtubule
Microtubules are polymers of tubulin that form part of the cytoskeleton and provide structure and shape to eukaryotic cells. Microtubules can be as long as 50 micrometres, as wide as 23 to 27 nm and have an inner diameter between 11 and 15 nm. They are formed by the polymerization of a dimer of two globular proteins, alpha and beta tubulin into protofilaments that can then associate laterally to form a hollow tube, the microtubule. The most common form of a microtubule consists of 13 protofilaments in the tubular arrangement. Microtubules play an important role in a number of cellular processes. They are involved in maintaining the structure of the cell and, together with microfilaments and intermediate filaments, they form the cytoskeleton. They also make up the internal structure of cilia and flagella. They provide platforms for intracellular transport and are involved in a variety of cellular processes, including the movement of secretory vesicles, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Kinesin
A kinesin is a protein belonging to a class of motor proteins found in eukaryotic cells. Kinesins move along microtubule (MT) filaments and are powered by the hydrolysis of adenosine triphosphate (ATP) (thus kinesins are ATPases, a type of enzyme). The active movement of kinesins supports several cellular functions including mitosis, meiosis and transport of cellular cargo, such as in axonal transport, and intraflagellar transport. Most kinesins walk towards the plus end of a microtubule, which, in most cells, entails transporting cargo such as protein and membrane components from the center of the cell towards the periphery. This form of transport is known as anterograde transport. In contrast, dyneins are motor proteins that move toward the minus end of a microtubule in retrograde transport. Discovery Kinesins were discovered in 1985, based on their motility in cytoplasm extruded from the giant axon of the squid. They turned out as MT-based anterograde intracellular tra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |