|
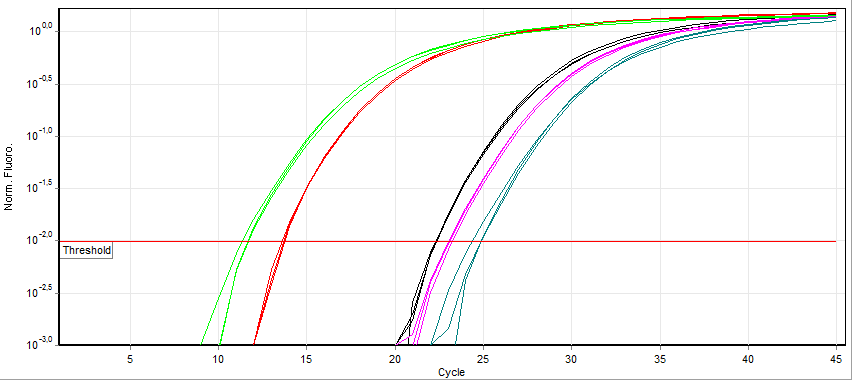
RT-qPCR
A real-time polymerase chain reaction (real-time PCR, or qPCR) is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR (i.e., in real time), not at its end, as in conventional PCR. Real-time PCR can be used quantitatively (quantitative real-time PCR) and semi-quantitatively (i.e., above/below a certain amount of DNA molecules) (semi-quantitative real-time PCR). Two common methods for the detection of PCR products in real-time PCR are (1) non-specific fluorescent dyes that intercalate with any double-stranded DNA and (2) sequence-specific DNA probes consisting of oligonucleotides that are labelled with a fluorescent reporter, which permits detection only after hybridization of the probe with its complementary sequence. The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines propose that the abbreviation ''qPCR'' be used for q ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Complementary DNA
In genetics, complementary DNA (cDNA) is DNA synthesized from a single-stranded RNA (e.g., messenger RNA ( mRNA) or microRNA (miRNA)) template in a reaction catalyzed by the enzyme reverse transcriptase. cDNA is often used to express a specific protein in a cell that does not normally express that protein (i.e., heterologous expression), or to sequence or quantify mRNA molecules using DNA based methods (qPCR, RNA-seq). cDNA that codes for a specific protein can be transferred to a recipient cell for expression, often bacterial or yeast expression systems. cDNA is also generated to analyze transcriptomic profiles in bulk tissue, single cells, or single nuclei in assays such as microarrays, qPCR, and RNA-seq. cDNA is also produced naturally by retroviruses (such as HIV-1, HIV-2, simian immunodeficiency virus, etc.) and then integrated into the host's genome, where it creates a provirus. The term ''cDNA'' is also used, typically in a bioinformatics context, to refer ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reverse Transcription Polymerase Chain Reaction
Reverse transcription polymerase chain reaction (RT-PCR) is a laboratory technique combining reverse transcription of RNA into DNA (in this context called complementary DNA or cDNA) and amplification of specific DNA targets using polymerase chain reaction (PCR). It is primarily used to measure the amount of a specific RNA. This is achieved by monitoring the amplification reaction using fluorescence, a technique called real-time PCR or quantitative PCR (qPCR). Combined RT-PCR and qPCR are routinely used for analysis of gene expression and quantification of viral RNA in research and clinical settings. The close association between RT-PCR and qPCR has led to metonymic use of the term qPCR to mean RT-PCR. Such use may be confusing, as RT-PCR can be used without qPCR, for example to enable molecular cloning, sequencing or simple detection of RNA. Conversely, qPCR may be used without RT-PCR, for example to quantify the copy number of a specific piece of DNA. Nomenclature The combin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is a method widely used to rapidly make millions to billions of copies (complete or partial) of a specific DNA sample, allowing scientists to take a very small sample of DNA and amplify it (or a part of it) to a large enough amount to study in detail. PCR was invented in 1983 by the American biochemist Kary Mullis at Cetus Corporation; Mullis and biochemist Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993. PCR is fundamental to many of the procedures used in genetic testing and research, including analysis of ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory research for a broad variety of applications including biomedical research ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MIQE
The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines are a set of protocols for conducting and reporting quantitative real-time PCR experiments and data, as devised by Bustin et al. in 2009. They were devised after a paper was published in 2002 that claimed to detect measles virus in children with autism through the use of RT-qPCR, but the results proved to be completely unreproducible by other scientists. The authors themselves also did not try to reproduce them and the raw data was found to have a large amount of errors and basic mistakes in analysis. This incident prompted Stephen Bustin to create the MIQE guidelines to provide a baseline level of quality for qPCR data published in scientific literature. Purpose The MIQE guidelines were created due to the low quality of qPCR data submitted to academic journals at the time, which was only becoming more common as Next Generation Sequencing machinery allowed for such experiments to be ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication. The process of gene expression is used by all known life— eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea), and utilized by viruses—to generate the macromolecular machinery for life. In genetics, gene expression is the most fundamental level at which the genotype gives rise to the phen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sensor
A sensor is a device that produces an output signal for the purpose of sensing a physical phenomenon. In the broadest definition, a sensor is a device, module, machine, or subsystem that detects events or changes in its environment and sends the information to other electronics, frequently a computer processor. Sensors are always used with other electronics. Sensors are used in everyday objects such as touch-sensitive elevator buttons ( tactile sensor) and lamps which dim or brighten by touching the base, and in innumerable applications of which most people are never aware. With advances in micromachinery and easy-to-use microcontroller platforms, the uses of sensors have expanded beyond the traditional fields of temperature, pressure and flow measurement, for example into MARG sensors. Analog sensors such as potentiometers and force-sensing resistors are still widely used. Their applications include manufacturing and machinery, airplanes and aerospace, cars, medicine, r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
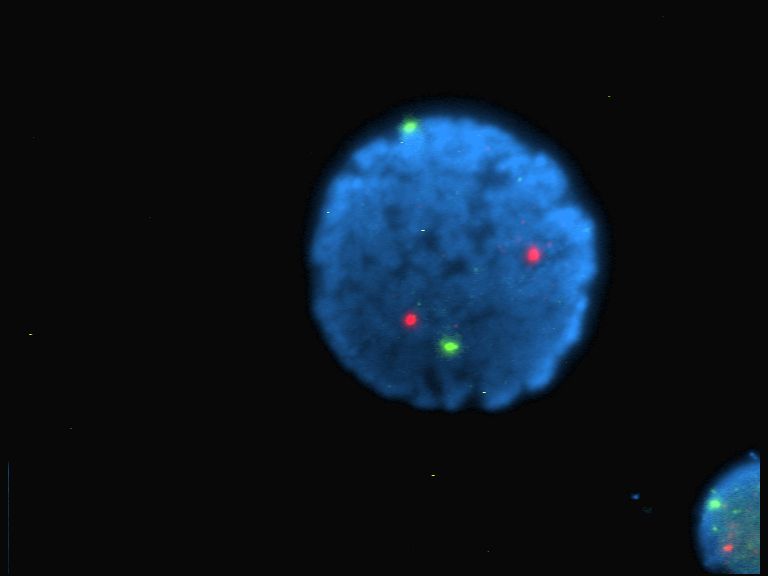
Fluorophore
A fluorophore (or fluorochrome, similarly to a chromophore) is a fluorescent chemical compound that can re-emit light upon light excitation. Fluorophores typically contain several combined aromatic groups, or planar or cyclic molecules with several π bonds. Fluorophores are sometimes used alone, as a tracer in fluids, as a dye for staining of certain structures, as a substrate of enzymes, or as a probe or indicator (when its fluorescence is affected by environmental aspects such as polarity or ions). More generally they are covalently bonded to a macromolecule, serving as a marker (or dye, or tag, or reporter) for affine or bioactive reagents ( antibodies, peptides, nucleic acids). Fluorophores are notably used to stain tissues, cells, or materials in a variety of analytical methods, i.e., fluorescent imaging and spectroscopy. Fluorescein, via its amine-reactive isothiocyanate derivative fluorescein isothiocyanate (FITC), has been one of the most popular fluorophor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Thermal Cycler
The thermal cycler (also known as a thermocycler, PCR machine or DNA amplifier) is a laboratory apparatus most commonly used to amplify segments of DNA via the polymerase chain reaction (PCR). Thermal cyclers may also be used in laboratories to facilitate other temperature-sensitive reactions, including restriction enzyme digestion or rapid diagnostics. The device has a ''thermal block'' with holes where tubes holding the reaction mixtures can be inserted. The cycler then raises and lowers the temperature of the block in discrete, pre-programmed steps. History The earliest thermal cyclers were designed for use with the Klenow fragment of DNA polymerase I. Since this enzyme is destroyed during each heating step of the amplification process, new enzyme had to be added every cycle. This led to a cumbersome machine based on an automated pipettor, with open reaction tubes. Later, the PCR process was adapted to the use of thermostable DNA polymerase from '' Thermus aquaticus' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Wavelength
In physics, the wavelength is the spatial period of a periodic wave—the distance over which the wave's shape repeats. It is the distance between consecutive corresponding points of the same phase on the wave, such as two adjacent crests, troughs, or zero crossings, and is a characteristic of both traveling waves and standing waves, as well as other spatial wave patterns. The inverse of the wavelength is called the spatial frequency. Wavelength is commonly designated by the Greek letter '' lambda'' (λ). The term ''wavelength'' is also sometimes applied to modulated waves, and to the sinusoidal envelopes of modulated waves or waves formed by interference of several sinusoids. Assuming a sinusoidal wave moving at a fixed wave speed, wavelength is inversely proportional to frequency of the wave: waves with higher frequencies have shorter wavelengths, and lower frequencies have longer wavelengths. Wavelength depends on the medium (for example, vacuum, air, or water) that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Buffer Solution
A buffer solution (more precisely, pH buffer or hydrogen ion buffer) is an aqueous solution consisting of a mixture of a weak acid and its conjugate base, or vice versa. Its pH changes very little when a small amount of strong acid or base is added to it. Buffer solutions are used as a means of keeping pH at a nearly constant value in a wide variety of chemical applications. In nature, there are many living systems that use buffering for pH regulation. For example, the bicarbonate buffering system is used to regulate the pH of blood, and bicarbonate also acts as a buffer in the ocean. Principles of buffering Buffer solutions resist pH change because of a chemical equilibrium between the weak acid HA and its conjugate base A−: When some strong acid is added to an equilibrium mixture of the weak acid and its conjugate base, hydrogen ions (H+) are added, and the equilibrium is shifted to the left, in accordance with Le Chatelier's principle. Because of this, the hydroge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Microarray
A DNA microarray (also commonly known as DNA chip or biochip) is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome. Each DNA spot contains picomoles (10−12 moles) of a specific DNA sequence, known as '' probes'' (or ''reporters'' or '' oligos''). These can be a short section of a gene or other DNA element that are used to hybridize a cDNA or cRNA (also called anti-sense RNA) sample (called ''target'') under high-stringency conditions. Probe-target hybridization is usually detected and quantified by detection of fluorophore-, silver-, or chemiluminescence-labeled targets to determine relative abundance of nucleic acid sequences in the target. The original nucleic acid arrays were macro arrays approximately 9 cm × 12 cm and the first computerized image based analysis was published in 1981. It was i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |