|
RNA-sequencing
RNA-Seq (named as an abbreviation of RNA sequencing) is a technique that uses next-generation sequencing to reveal the presence and quantity of RNA molecules in a biological sample, providing a snapshot of gene expression in the sample, also known as transcriptome. RNA-Seq facilitates the ability to look at alternative gene spliced transcripts, post-transcriptional modifications, gene fusion, mutations/SNPs and changes in gene expression over time, or differences in gene expression in different groups or treatments. In addition to mRNA transcripts, RNA-Seq can look at different populations of RNA to include total RNA, small RNA, such as miRNA, tRNA, and ribosomal profiling. RNA-Seq can also be used to determine exon/intron boundaries and verify or amend previously annotated 5' and 3' gene boundaries. Recent advances in RNA-Seq include single cell sequencing, bulk RNA sequencing, 3' mRNA-sequencing, in situ sequencing of fixed tissue, and native RNA molecule sequencin g with ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-cell Transcriptomics
Single-cell transcriptomics examines the gene expression level of individual Cell (biology), cells in a given population by simultaneously measuring the RNA concentration (conventionally only messenger RNA (mRNA)) of hundreds to thousands of genes. Single-cell transcriptomics makes it possible to unravel Heterogenous, heterogeneous cell populations, reconstruct cellular developmental pathways, and model transcriptional dynamics — all previously masked in bulk RNA sequencing. Background The development of high-throughput RNA sequencing (RNA-seq) and microarrays has made gene expression analysis a routine. RNA analysis was previously limited to tracing individual Transcription (biology), transcripts by Northern blots or quantitative PCR. Higher throughput and speed allow researchers to frequently characterize the expression profiles of populations of thousands of cells. The data from bulk assays has led to identifying genes differentially expressed in distinct cell populations, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
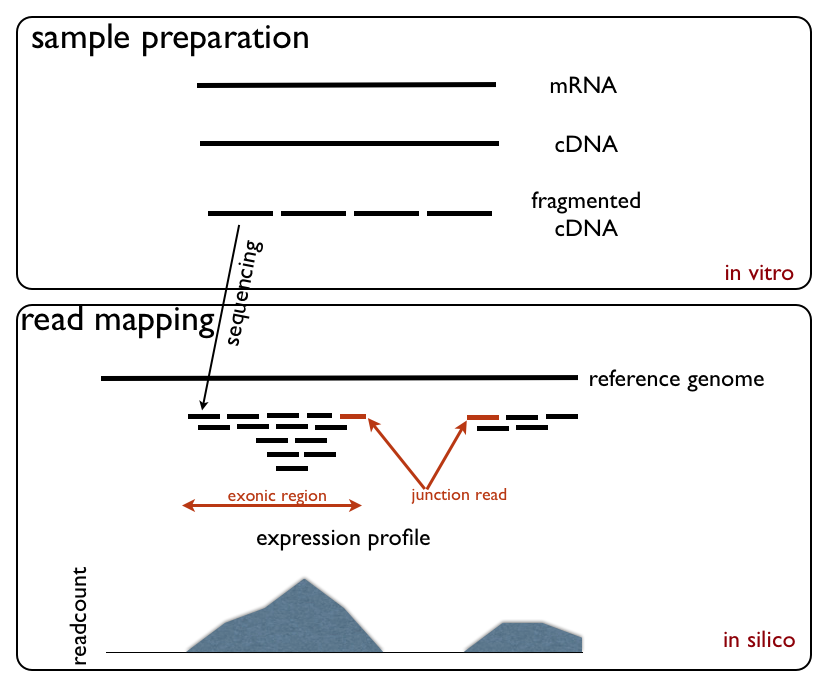
Transcriptomics
Transcriptomics technologies are the techniques used to study an organism's transcriptome, the sum of all of its RNA, RNA transcripts. The information content of an organism is recorded in the DNA of its genome and Gene expression, expressed through Transcription (genetics), transcription. Here, mRNA serves as a transient intermediary molecule in the information network, whilst non-coding RNAs perform additional diverse functions. A transcriptome captures a snapshot in time of the total transcripts present in a cell (biology), cell. Transcriptomics technologies provide a broad account of which cellular processes are active and which are dormant. A major challenge in molecular biology is to understand how a single genome gives rise to a variety of cells. Another is how gene expression is regulated. The first attempts to study whole transcriptomes began in the early 1990s. Subsequent technological advances since the late 1990s have repeatedly transformed the field and made transcript ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNAseq Over Time (Pubmed)
RNA-Seq (named as an abbreviation of RNA sequencing) is a technique that uses next-generation sequencing to reveal the presence and quantity of RNA molecules in a biological sample, providing a snapshot of gene expression in the sample, also known as transcriptome. RNA-Seq facilitates the ability to look at alternative gene spliced transcripts, post-transcriptional modifications, gene fusion, mutations/SNPs and changes in gene expression over time, or differences in gene expression in different groups or treatments. In addition to mRNA transcripts, RNA-Seq can look at different populations of RNA to include total RNA, small RNA, such as miRNA, tRNA, and ribosomal profiling. RNA-Seq can also be used to determine exon/intron boundaries and verify or amend previously annotated 5' and 3' gene boundaries. Recent advances in RNA-Seq include single cell sequencing, bulk RNA sequencing, 3' mRNA-sequencing, in situ sequencing of fixed tissue, and native RNA molecule sequencin g with ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alternative Splicing
Alternative splicing, alternative RNA splicing, or differential splicing, is an alternative RNA splicing, splicing process during gene expression that allows a single gene to produce different splice variants. For example, some exons of a gene may be included within or excluded from the final RNA product of the gene. This means the exons are joined in different combinations, leading to different splice variants. In the case of protein-coding genes, the proteins translated from these splice variants may contain differences in their amino acid sequence and in their biological functions (see Figure). Biologically relevant alternative splicing occurs as a normal phenomenon in eukaryotes, where it increases the number of proteins that can be encoded by the genome. In humans, it is widely believed that ~95% of multi-exonic genes are alternatively spliced to produce functional alternative products from the same gene but many scientists believe that most of the observed splice variants ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Summary Of RNA-Seq
Summary may refer to: * Abstract (summary), shortening a passage or a write-up without changing its meaning but by using different words and sentences * Epitome, a summary or miniature form * Abridgement, the act of reducing a written work into a shorter form * Summary or executive summary of a document, a short document or section that summarizes a longer document such as a report or proposal or a group of related reports * Introduction (writing) * Summary (law), which has several meanings in law * Automatic summarization, the use of a computer program to produce an abstract or abridgement See also * Overview (other) * Recap (other) * Synopsis (other) {{disambiguation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serial Analysis Of Gene Expression
Serial Analysis of Gene Expression (SAGE) is a transcriptomic technique used by molecular biologists to produce a snapshot of the messenger RNA population in a sample of interest in the form of small tags that correspond to fragments of those transcripts. Several variants have been developed since, most notably a more robust version, LongSAGE, RL-SAGE and the most recent SuperSAGE. Many of these have improved the technique with the capture of longer tags, enabling more confident identification of a source gene. Overview Briefly, SAGE experiments proceed as follows: # The mRNA of an input sample (e.g. a tumour) is isolated and a reverse transcriptase and biotinylated primers are used to synthesize cDNA from mRNA. # The cDNA is bound to Streptavidin beads via interaction with the biotin attached to the primers, and is then cleaved using a restriction endonuclease called an anchoring enzyme (AE). The location of the cleavage site and thus the length of the remaining cDNA bound ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
A Priori And A Posteriori
('from the earlier') and ('from the later') are Latin phrases used in philosophy to distinguish types of knowledge, justification, or argument by their reliance on experience. knowledge is independent from any experience. Examples include mathematics,Some associationist philosophers have contended that mathematics comes from experience and is not a form of any ''a priori'' knowledge () tautologies and deduction from pure reason. Galen Strawson has stated that an argument is one in which "you can see that it is true just lying on your couch. You don't have to get up off your couch and go outside and examine the way things are in the physical world. You don't have to do any science." () knowledge depends on empirical evidence. Examples include most fields of science and aspects of personal knowledge. The terms originate from the analytic methods found in '' Organon'', a collection of works by Aristotle. Prior analytics () is about deductive logic, which comes from ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sanger Sequencing
Sanger sequencing is a method of DNA sequencing that involves electrophoresis and is based on the random incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication. After first being developed by Frederick Sanger and colleagues in 1977, it became the most widely used sequencing method for approximately 40 years. An automated instrument using slab gel electrophoresis and fluorescent labels was first commercialized by Applied Biosystems in March 1987. Later, automated slab gels were replaced with automated capillary array electrophoresis. Recently, higher volume Sanger sequencing has been replaced by next generation sequencing methods, especially for large-scale, automated genome analyses. However, the Sanger method remains in wide use for smaller-scale projects and for validation of deep sequencing results. It still has the advantage over short-read sequencing technologies (like Illumina) in that it can produce DNA sequence reads of > ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Expressed Sequence Tag
In genetics, an expressed sequence tag (EST) is a short sub-sequence of a cDNA sequence. ESTs may be used to identify gene transcripts, and were instrumental in gene discovery and in gene-sequence determination. The identification of ESTs has proceeded rapidly, with approximately 74.2 million ESTs now available in public databases (e.g. GenBank 1 January 2013, all species). EST approaches have largely been superseded by whole genome and transcriptome sequencing and metagenome sequencing. An EST results from one-shot sequencing of a cloned cDNA. The cDNAs used for EST generation are typically individual clones from a cDNA library. The resulting sequence is a relatively low-quality fragment whose length is limited by current technology to approximately 500 to 800 nucleotides. Because these clones consist of DNA that is complementary to mRNA, the ESTs represent portions of expressed genes. They may be represented in databases as either cDNA/mRNA sequence or as the reverse compleme ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Complementary DNA
In genetics, complementary DNA (cDNA) is DNA that was reverse transcribed (via reverse transcriptase) from an RNA (e.g., messenger RNA or microRNA). cDNA exists in both single-stranded and double-stranded forms and in both natural and engineered forms. In engineered forms, it often is a copy (replicate) of the naturally occurring DNA from any particular organism's natural genome; the organism's own mRNA was naturally transcribed from its DNA, and the cDNA is reverse transcribed from the mRNA, yielding a duplicate of the original DNA. Engineered cDNA is often used to express a specific protein in a cell that does not normally express that protein (i.e., heterologous expression), or to sequence or quantify mRNA molecules using DNA based methods (qPCR, RNA-seq). cDNA that codes for a specific protein can be transferred to a recipient cell for expression as part of recombinant DNA, often bacterial or yeast expression systems. cDNA is also generated to analyze transcriptomic pr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Next-gen Sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, thymine, cytosine, and guanine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery. Knowledge of DNA sequences has become indispensable for basic biological research, DNA Genographic Projects and in numerous applied fields such as medical diagnosis, biotechnology, forensic biology, virology and biological systematics. Comparing healthy and mutated DNA sequences can diagnose different diseases including various cancers, characterize antibody repertoire, and can be used to guide patient treatment. Having a quick way to sequence DNA allows for faster and more individualized medical care to be administered, and for more organisms to be identified and cataloged. The rapid advancements in DNA sequencing technology ha ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Prostate Cancer
Prostate cancer is the neoplasm, uncontrolled growth of cells in the prostate, a gland in the male reproductive system below the bladder. Abnormal growth of the prostate tissue is usually detected through Screening (medicine), screening tests, typically blood tests that check for prostate-specific antigen (PSA) levels. Those with high levels of PSA in their blood are at increased risk for developing prostate cancer. Diagnosis requires a prostate biopsy, biopsy of the prostate. If cancer is present, the pathologist assigns a Gleason score; a higher score represents a more dangerous tumor. Medical imaging is performed to look for cancer that has spread outside the prostate. Based on the Gleason score, PSA levels, and imaging results, a cancer case is assigned a cancer staging, stage 1 to 4. A higher stage signifies a more advanced, more dangerous disease. Most prostate tumors remain small and cause no health problems. These are managed with active surveillance of prostate cancer, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |