|
Protein Topology
Protein topology is a property of protein molecule that does not change under deformation (without cutting or breaking a bond). Frameworks Two main topology frameworks have been developed and applied to protein molecules. Knot Theory Knot theory which categorises chain entanglements. The usage of knot theory is limited to a small percentage of proteins as most of them are unknot. Circuit topology Circuit topology categorises intra-chain contacts based on their arrangements. Other Uses In biology literature, the term topology is also used to refer to mutual orientation of regular secondary structures, such as alpha-helices and beta strands in protein structure For example, two adjacent interacting alpha-helices or beta-strands can go in the same or in opposite directions. Topology diagrams of dif ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Anthrax Toxin Protein Key Motif
Anthrax is an infection caused by the bacterium '' Bacillus anthracis''. It can occur in four forms: skin, lungs, intestinal, and injection. Symptom onset occurs between one day and more than two months after the infection is contracted. The skin form presents with a small blister with surrounding swelling that often turns into a painless ulcer with a black center. The inhalation form presents with fever, chest pain and shortness of breath. The intestinal form presents with diarrhea (which may contain blood), abdominal pains, nausea and vomiting. The injection form presents with fever and an abscess at the site of drug injection. According to the USA's Centers for Disease Control and Prevention, the first clinical descriptions of cutaneous anthrax were given by Maret in 1752 and Fournier in 1769. Before that anthrax had been described only through historical accounts. The Prussian scientist Robert Koch (1843–1910) was the first to identify ''Bacillus anthracis'' as the bacte ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Knot Theory
In the mathematical field of topology, knot theory is the study of knot (mathematics), mathematical knots. While inspired by knots which appear in daily life, such as those in shoelaces and rope, a mathematical knot differs in that the ends are joined so it cannot be undone, Unknot, the simplest knot being a ring (or "unknot"). In mathematical language, a knot is an embedding of a circle in 3-dimensional Euclidean space, \mathbb^3 (in topology, a circle is not bound to the classical geometric concept, but to all of its homeomorphisms). Two mathematical knots are equivalent if one can be transformed into the other via a deformation of \mathbb^3 upon itself (known as an ambient isotopy); these transformations correspond to manipulations of a knotted string that do not involve cutting it or passing through itself. Knots can be described in various ways. Using different description methods, there may be more than one description of the same knot. For example, a common method of descr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Circuit Topology
The circuit topology of a folded linear polymer refers to the arrangement of its intra-molecular contacts. Examples of linear polymers with intra-molecular contacts are nucleic acids and proteins. Proteins fold via formation of contacts of various nature, including hydrogen bonds, disulfide bonds, and beta-beta interactions. Contacts in the genome are established via protein bridges including CTCF and cohesins and are measured by technologies including Hi-C. Circuit topology categorises the topological arrangement of these physical contacts, that are referred to as hard contacts. Furthermore, chains can fold via knotting (or formation of "soft" contacts). Circuit topology uses a similar language to categorise both "soft" and "hard" contacts, and provides a full description of a folded linear chain. A simple example of a folded chain is a chain with two hard contacts. For a chain with two binary contacts, three arrangements are available: parallel, series and crossed. For a chain ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Secondary Structure
Protein secondary structure is the three dimensional form of ''local segments'' of proteins. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein folds into its three dimensional tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the amino hydrogen and carboxyl oxygen atoms in the peptide backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik Linderstrøm-Lang at Stanford in 1952. Other types of biopolymers such as nucleic acids also possess characteristic secondary structures. Types The most common secondary structures ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha-helices
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence. The alpha helix is also called a classic Pauling–Corey–Branson α-helix. The name 3.613-helix is also used for this type of helix, denoting the average number of residues per helical turn, with 13 atoms being involved in the ring formed by the hydrogen bond. Among types of local structure in proteins, the α-helix is the most extreme and the most predictable from sequence, as well as the most prevalent. Discovery In the early 1930s, William Astbury showed that there were drastic changes in the X-ray fiber diffraction of moist wool or hair fibers upon significant stretching. The data suggested that the unstretched fibers had a coiled molecular structure with a characteristic repeat of ≈. Astbur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta Strand
The beta sheet, (β-sheet) (also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, notably Alzheimer's disease. History The first β-sheet structure was proposed by William Astbury in the 1930s. He proposed the idea of hydrogen bonding between the peptide bonds of parallel or antiparallel extended β-strands. However, Astbury did not have the necessary data on the bond geometry of the amino acids in order to build accurate models, especially since he did not then know that the peptide bond was planar. A refined version was p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Structure
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, the monomers of the polymer. A single amino acid monomer may also be called a ''residue'' indicating a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reactions, in which the amino acids lose one water molecule per reaction in order to attach to one another with a peptide bond. By convention, a chain under 30 amino acids is often identified as a peptide, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their three-dimensional structure. This is t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PDBsum
PDBsum is a database that provides an overview of the contents of each 3D macromolecular structure deposited in the Protein Data Bank. The original version of the database was developed around 1995 by Roman Laskowski and collaborators at University College London. As of 2014, PDBsum is maintained by Laskowski and collaborators in the laboratory of Janet Thornton at the European Bioinformatics Institute (EBI). Each structure in the PDBsum database includes an image of structure (main view, Bottom view and right view), molecular components contained in the complex(structure), enzyme reaction diagram if appropriate, Gene ontology functional assignments, a 1D sequence annotated by Pfam and InterPro domain assignments, description of bound molecules and graphic showing interactions between protein and secondary structure, schematic diagrams of protein–protein interactions, analysis of clefts contained within the structure and links to external databases. The RasMol and Jmol molecul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
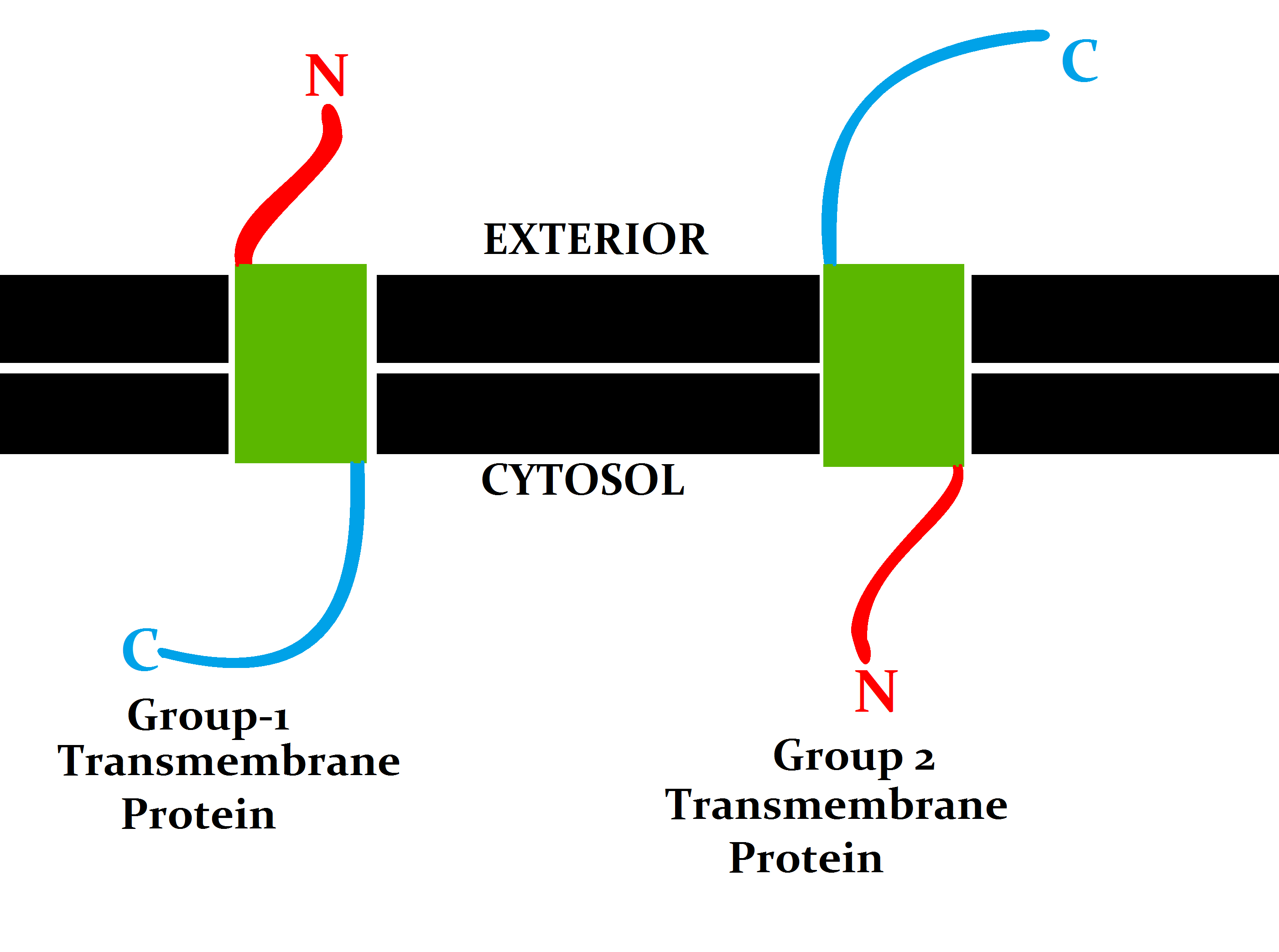
Membrane Topology
Topology of a transmembrane protein refers to locations of N- and C-termini of membrane-spanning polypeptide chain with respect to the inner or outer sides of the biological membrane occupied by the protein. Several databases provide experimentally determined topologies of membrane proteins. They include Uniprot, TOPDB, OPM, and ExTopoDB. There is also a database of domains located conservatively on a certain side of membranes, TOPDOM. Several computational methods were developed, with a limited success, for predicting transmembrane alpha-helices and their topology. Pioneer methods utilized the fact that membrane-spanning regions contain more hydrophobic residues than other parts of the protein, however applying different hydrophobic scales altered the prediction results. Later, several statistical methods were developed to improve the topography prediction and a special alignment method was introduced. According to the positive-inside rule, cytosolic loops near the lipid bilaye ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Folding
Protein folding is the physical process by which a protein chain is translated to its native three-dimensional structure, typically a "folded" conformation by which the protein becomes biologically functional. Via an expeditious and reproducible process, a polypeptide folds into its characteristic three-dimensional structure from a random coil. Each protein exists first as an unfolded polypeptide or random coil after being translated from a sequence of mRNA to a linear chain of amino acids. At this stage the polypeptide lacks any stable (long-lasting) three-dimensional structure (the left hand side of the first figure). As the polypeptide chain is being synthesized by a ribosome, the linear chain begins to fold into its three-dimensional structure. Folding of many proteins begins even during translation of the polypeptide chain. Amino acids interact with each other to produce a well-defined three-dimensional structure, the folded protein (the right hand side of the figure), ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Structure
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, the monomers of the polymer. A single amino acid monomer may also be called a ''residue'' indicating a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reactions, in which the amino acids lose one water molecule per reaction in order to attach to one another with a peptide bond. By convention, a chain under 30 amino acids is often identified as a peptide, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their three-dimensional structure. This is t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |