|
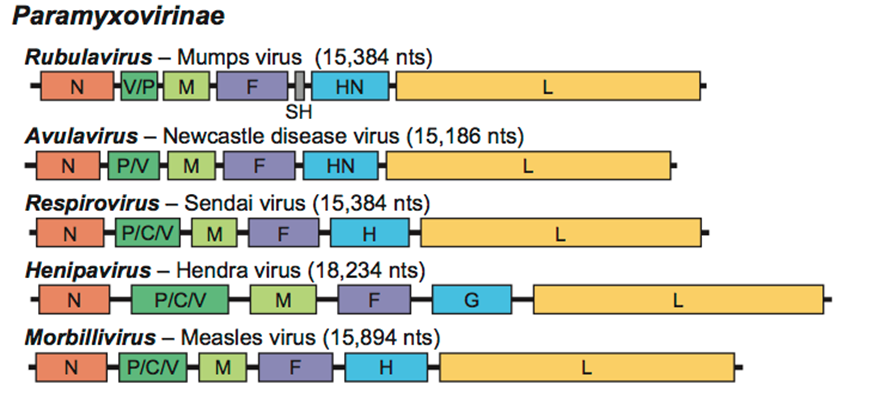
Paramyxoviridae
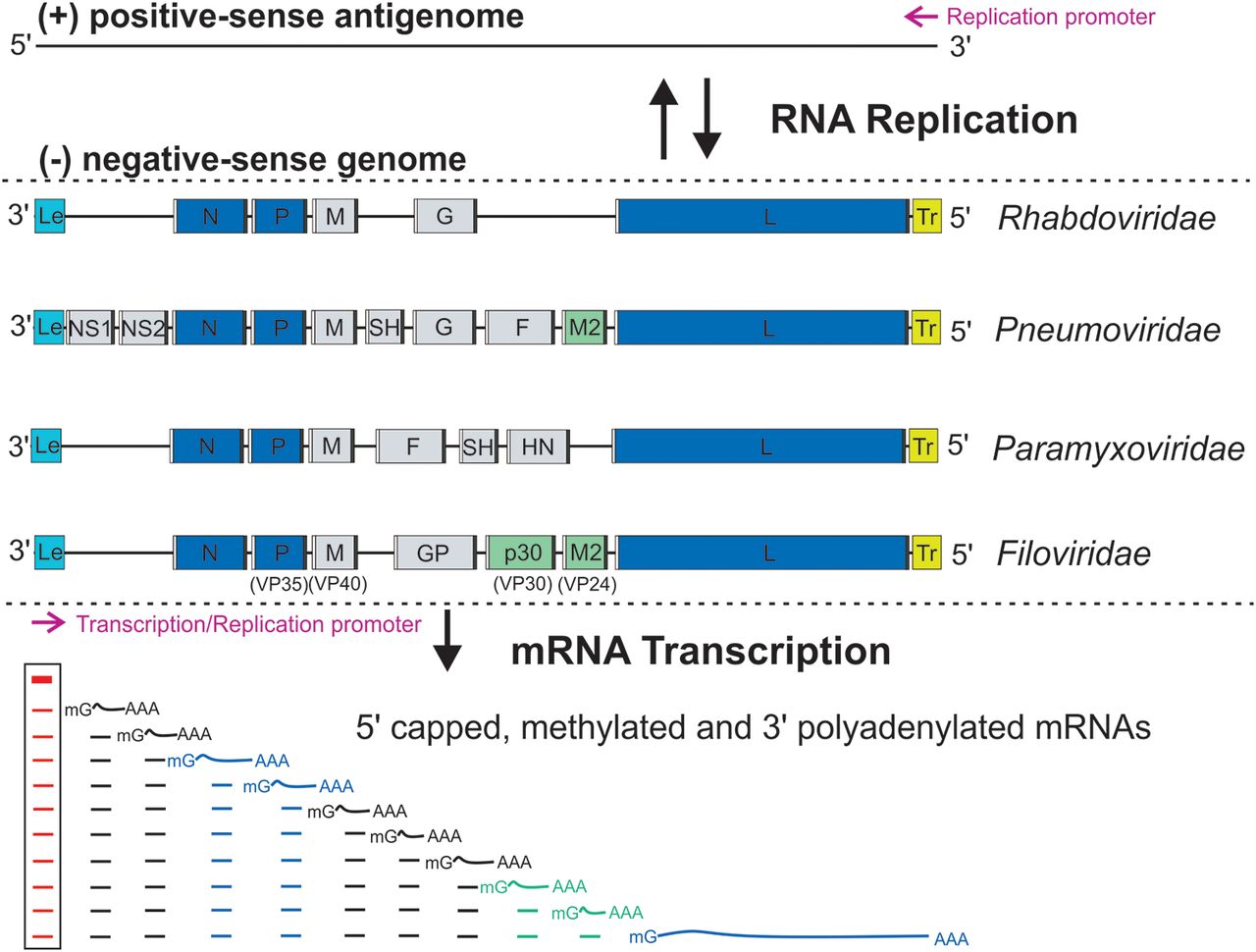
''Paramyxoviridae'' (from Ancient Greek, Greek ''para-'' “by the side of” and ''myxa'' “mucus”) is a family of negative-strand RNA viruses in the order ''Mononegavirales''. Vertebrates serve as natural hosts. Diseases associated with this family include measles, mumps, and respiratory tract infections. The family has four subfamilies, 17 genera, and 78 species, three genera of which are unassigned to a subfamily. Structure Virions are enveloped and can be spherical or pleomorphic and capable of producing filamentous virions. The diameter is around 150 nm. Genomes are linear, around 15kb in length. Fusion proteins and attachment proteins appear as spikes on the virion surface. Matrix proteins inside the envelope stabilise virus structure. The nucleocapsid core is composed of the genomic RNA, nucleocapsid proteins, phosphoproteins and polymerase proteins. Genome The genome is nonsegmented, negative-sense RNA, 15–19 kilobases in length, and contains six to 10 gene ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Avulavirinae
''Avulavirinae'' is a subfamily of viruses in the family ''Paramyxoviridae''. Members of the subfamily are collectively known as avulaviruses. All members of the subfamily primarily infect birds. ''Avulavirinae'' was previously recognized as the genus ''Avulavirus'' before being elevated to a subfamily. The term ''avula'' comes from "avian rubula", distinguishing it from rubulaviruses of the subfamily ''Rubulavirinae'' due to avulaviruses only infecting birds and translating protein V from an edited RNA transcript. The most notable avulavirus is the Newcastle disease, Newcastle disease virus, a strain of ''Avian orthoavulavirus 1''. Characteristics Like all Paramyxoviruses, avulaviruses are Enveloped virus, enveloped Negative-strand RNA virus, negative-strand RNA viruses. Avulaviruses have a hemagglutinin-neuraminidase attachment protein and do not produce a non-structural protein C. Avulaviruses can be separated into distinct serotypes using hemagglutination assay and neuraminidas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Negative-strand RNA Virus
Negative-strand RNA viruses (−ssRNA viruses) are a group of related viruses that have negative-sense, single-stranded genomes made of ribonucleic acid. They have genomes that act as complementary strands from which messenger RNA (mRNA) is synthesized by the viral enzyme RNA-dependent RNA polymerase (RdRp). During replication of the viral genome, RdRp synthesizes a positive-sense antigenome that it uses as a template to create genomic negative-sense RNA. Negative-strand RNA viruses also share a number of other characteristics: most contain a viral envelope that surrounds the capsid, which encases the viral genome, −ssRNA virus genomes are usually linear, and it is common for their genome to be segmented. Negative-strand RNA viruses constitute the phylum ''Negarnaviricota'', in the kingdom '' Orthornavirae'' and realm '' Riboviria''. They are descended from a common ancestor that was a double-stranded RNA (dsRNA) virus, and they are considered to be a sister clade of reovi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rubulavirinae
''Rubulavirinae'' is a subfamily of viruses in the family ''Paramyxoviridae''. Humans, apes, pigs, and dogs serve as natural hosts. There are currently 18 species in the two genera ''Orthorubulavirus'' and ''Pararubulavirus''. Diseases associated with this genus include mumps. Members of the subfamily are collectively called rubulaviruses. The subfamily was previously a genus named ''Rubulavirus'' but was elevated to subfamily in 2018. Viruses of this subfamily appear to be most closely related to members of ''Avulavirinae''. Taxonomy Subfamily: ''Rubulavirinae'' :Genus: ''Orthorubulavirus'' :Genus: ''Pararubulavirus'' Structure Rubulavirions are enveloped, with spherical geometries. The diameter is around 150 nm. Rubulavirus genomes are linear, around 15kb in length. The genome codes for 8 proteins. Life cycle Viral replication is cytoplasmic. Entry into the host cells is achieved after viral attachment to host cells. Replication follows the negative stranded RNA virus r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Orthoparamyxovirinae
''Orthoparamyxovirinae'' is a subfamily of viruses in the family ''Paramyxoviridae''. Most genera in the subfamily belonged to a previous subfamily, ''Paramyxovirinae'', which was abolished in 2015. The current subfamily, with those genera, was established in 2018. Taxonomy *''Aquaparamyxovirus'' *'' Ferlavirus'' *''Henipavirus'' *'' Jeilongvirus'' *''Morbillivirus'' *'' Narmovirus'' *''Respirovirus ''Respirovirus'' is a genus of viruses in the order ''Mononegavirales'', in the family ''Paramyxoviridae''. Rodents and human serve as natural hosts. There are seven species in this genus. Diseases associated with this genus include: croup and ot ...'' *'' Salemvirus'' References External link ICTV Report: ''Paramyxoviridae''{{Taxonbar, from=Q69855699 Virus subfamilies Paramyxoviridae ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mononegavirales
''Mononegavirales'' is an order of negative-strand RNA viruses which have nonsegmented genomes. Some common members of the order are Ebola virus, human respiratory syncytial virus, measles virus, mumps virus, Nipah virus, and rabies virus. All of these viruses cause significant disease in humans. Many other important pathogens of nonhuman animals and plants are also in the group. The order includes eleven virus families: '' Artoviridae'', ''Bornaviridae'', ''Filoviridae'', ''Lispiviridae'', ''Mymonaviridae'', ''Nyamiviridae'', ''Paramyxoviridae'', ''Pneumoviridae'', ''Rhabdoviridae'', '' Sunviridae'', and ''Xinmoviridae''. Use of term The order ''Mononegavirales'' (pronounced: ) According to the rules for taxon naming established by the International Committee on Taxonomy of Viruses (ICTV), the name ''Mononegavirales'' is always to be capitalized, italicized, and never abbreviated. The names of the order's physical members ("mononegaviruses" or "mononegavirads") are to be writte ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rule Of Six (viruses)
The rule of six is a feature of some paramyxovirus genomes. These RNA viruses have genes made from RNA and not DNA, and their whole genome – that is the number of nucleotides – is always a multiple of six. This is because during their replication, these viruses are dependent on nucleoprotein molecules A molecule is a group of two or more atoms held together by attractive forces known as chemical bonds; depending on context, the term may or may not include ions which satisfy this criterion. In quantum physics, organic chemistry, and bioche ... that each bind to six nucleotides.. References {{reflist Paramyxoviridae Genetics ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hemagglutinin-neuraminidase
Hemagglutinin-neuraminidase refers to a single viral protein that has both hemagglutinin and (endo) neuraminidase activity. This is in contrast to the proteins found in influenza, where both functions exist but in two separate proteins. Its neuraminidase domain has the CAZy designation glycoside hydrolase family 83 (GH83). It does show a structural similarity to influenza viral neuraminidase and has a six-bladed beta-propeller structure. This Pfam entry also matches measles hemagglutinin (cd15467), which has a "dead" neuraminidase part repurposed as a receptor binding site. Hemagglutinin-neuraminidase allows the virus to stick to a potential host cell, and cut itself loose if necessary. Hemagglutinin-neuraminidase can be found in a variety of paramyxoviruses including mumps virus, human parainfluenza virus 3, and the avian pathogen Newcastle disease virus. Types include: * Mumps hemagglutinin-neuraminidase * Parainfluenza hemagglutinin-neuraminidase Parainfluenza hemagglutin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Measles
Measles is a highly contagious infectious disease caused by measles virus. Symptoms usually develop 10–12 days after exposure to an infected person and last 7–10 days. Initial symptoms typically include fever, often greater than , cough, runny nose, and inflamed eyes. Small white spots known as Koplik's spots may form inside the mouth two or three days after the start of symptoms. A red, flat rash which usually starts on the face and then spreads to the rest of the body typically begins three to five days after the start of symptoms. Common complications include diarrhea (in 8% of cases), middle ear infection (7%), and pneumonia (6%). These occur in part due to measles-induced immunosuppression. Less commonly seizures, blindness, or inflammation of the brain may occur. Other names include ''morbilli'', ''rubeola'', ''red measles'', and ''English measles''. Both rubella, also known as ''German measles'', and roseola are different diseases caused by unrelated viruses. Mea ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA-dependent RNA Polymerase
RNA-dependent RNA polymerase (RdRp) or RNA replicase is an enzyme that catalyzes the replication of RNA from an RNA template. Specifically, it catalyzes synthesis of the RNA strand complementary to a given RNA template. This is in contrast to typical DNA-dependent RNA polymerases, which all organisms use to catalyze the transcription of RNA from a DNA template. RdRp is an essential protein encoded in the genomes of most RNA-containing viruses with no DNA stage including SARS-CoV-2. Some eukaryotes also contain RdRps, which are involved in RNA interference and differ structurally from viral RdRps. History Viral RdRps were discovered in the early 1960s from studies on mengovirus and polio virus when it was observed that these viruses were not sensitive to actinomycin D, a drug that inhibits cellular DNA-directed RNA synthesis. This lack of sensitivity suggested that there is a virus-specific enzyme that could copy RNA from an RNA template and not from a DNA template. Distr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Paramyxovirus Genome Structure '' was known as ''Paramyxovirus'' (with italics) until 1998
{{disambig ...
Paramyxovirus can refer to two different groups of viruses: *a member of the family ''Paramyxoviridae'' can be called a paramyxovirus (no italics) *the genus ''Respirovirus ''Respirovirus'' is a genus of viruses in the order ''Mononegavirales'', in the family ''Paramyxoviridae''. Rodents and human serve as natural hosts. There are seven species in this genus. Diseases associated with this genus include: croup and ot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hemagglutinin
In molecular biology, hemagglutinins (or ''haemagglutinin'' in British English) (from the Greek , 'blood' + Latin , 'glue') are receptor-binding membrane fusion glycoproteins produced by viruses in the ''Paramyxoviridae'' family. Hemagglutinins are responsible for binding to receptors on red blood cells to initiate viral attachment and infection. The agglutination of red cells occurs when antibodies on one cell bind to those on others, causing amorphous aggregates of clumped cells.Hemagglutinins recognize cell-surface glycoconjugates containing sialic acid on the surface of host red blood cells with a low affinity, and use them to enter the endosome of host cells. In the endosome, hemagglutinins are activated at a pH of 5 - 6.5, to undergo conformational changes that enable viral attachment through a fusion peptide. Agglutination and hemagglutinins were discovered by virologist George K. Hirst in 1941. Alfred Gottschalk proved in 1957 that hemagglutinins bind a virus to a host c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Viral Entry
Viral entry is the earliest stage of infection in the viral life cycle, as the virus comes into contact with the host cell (biology), cell and introduces viral material into the cell. The major steps involved in viral entry are shown below. Despite the variation among viruses, there are several shared generalities concerning viral entry. Reducing cellular proximity How a virus enters a cell is different depending on the type of virus it is. A virus with a Viral envelope, nonenveloped capsid enters the cell by attaching to the attachment factor located on a host cell. It then enters the cell by endocytosis or by making a hole in the membrane of the host cell and inserting its viral genome. Cell entry by enveloped viruses is more complicated. Enveloped viruses enter the cell by attaching to an attachment factor located on the surface of the host cell. They then enter by endocytosis or a direct membrane fusion event. The fusion event is when the virus membrane and the host cell m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |