|
Paleogenomics
Paleogenomics is a field of science based on the reconstruction and analysis of genomic information in extinct species. Improved methods for the extraction of ancient DNA (aDNA) from museum artifacts, ice cores, archeological or paleontological sites, and next-generation sequencing technologies have spurred this field. It is now possible to detect genetic drift, ancient population migration and interrelationships, the evolutionary history of extinct plant, animal and ''Homo'' species, and identification of phenotypic features across geographic regions. Scientists can also use paleogenomics to compare ancient ancestors against modern-day humans. The rising importance of paleogenomics is evident from the fact that the 2022 Nobel Prize in physiology or medicine was awarded to a Swedish geneticist Svante Pääbo 955- who worked on paleogenomics. Background Initially, aDNA sequencing involved cloning small fragments into bacteria, which proceeded with low efficiency due to the oxidati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Svante Pääbo
Svante Pääbo (; born 20 April 1955) is a Swedish geneticist who specialises in the field of evolutionary genetics. As one of the founders of paleogenetics, he has worked extensively on the Neanderthal genome. In 1997, he became founding director of the Department of Genetics at the Max Planck Institute for Evolutionary Anthropology in Leipzig, Germany. Since 1999, he has been an honorary professor at Leipzig University; he currently teaches molecular evolutionary biology at the university. He is also an adjunct professor at Okinawa Institute of Science and Technology, Japan. In 2022, he was awarded the Nobel Prize in Physiology or Medicine "for his discoveries concerning the genomes of extinct hominins and human evolution". Education and early life Pääbo was born in Stockholm, Sweden in 1955 and grew up there with his mother, Estonian chemist Karin Pääbo (1925–2013), who had escaped from the Soviet invasion in 1944 and arrived in Sweden as a refugee during World ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Species
In biology, a species is the basic unit of classification and a taxonomic rank of an organism, as well as a unit of biodiversity. A species is often defined as the largest group of organisms in which any two individuals of the appropriate sexes or mating types can produce fertile offspring, typically by sexual reproduction. Other ways of defining species include their karyotype, DNA sequence, morphology, behaviour or ecological niche. In addition, paleontologists use the concept of the chronospecies since fossil reproduction cannot be examined. The most recent rigorous estimate for the total number of species of eukaryotes is between 8 and 8.7 million. However, only about 14% of these had been described by 2011. All species (except viruses) are given a two-part name, a "binomial". The first part of a binomial is the genus to which the species belongs. The second part is called the specific name or the specific epithet (in botanical nomenclature, also sometimes i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
High Throughput Sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery. Knowledge of DNA sequences has become indispensable for basic biological research, DNA Genographic Projects and in numerous applied fields such as medical diagnosis, biotechnology, forensic biology, virology and biological systematics. Comparing healthy and mutated DNA sequences can diagnose different diseases including various cancers, characterize antibody repertoire, and can be used to guide patient treatment. Having a quick way to sequence DNA allows for faster and more individualized medical care to be administered, and for more organisms to be identified and cataloged. The rapid speed of sequencing attained with modern DNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Shotgun Sequencing
In genetics, shotgun sequencing is a method used for sequencing random DNA strands. It is named by analogy with the rapidly expanding, quasi-random shot grouping of a shotgun. The chain-termination method of DNA sequencing ("Sanger sequencing") can only be used for short DNA strands of 100 to 1000 base pairs. Due to this size limit, longer sequences are subdivided into smaller fragments that can be sequenced separately, and these sequences are assembled to give the overall sequence. In shotgun sequencing, DNA is broken up randomly into numerous small segments, which are sequenced using the chain termination method to obtain ''reads''. Multiple overlapping reads for the target DNA are obtained by performing several rounds of this fragmentation and sequencing. Computer programs then use the overlapping ends of different reads to assemble them into a continuous sequence. Shotgun sequencing was one of the precursor technologies that was responsible for enabling whole genome se ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
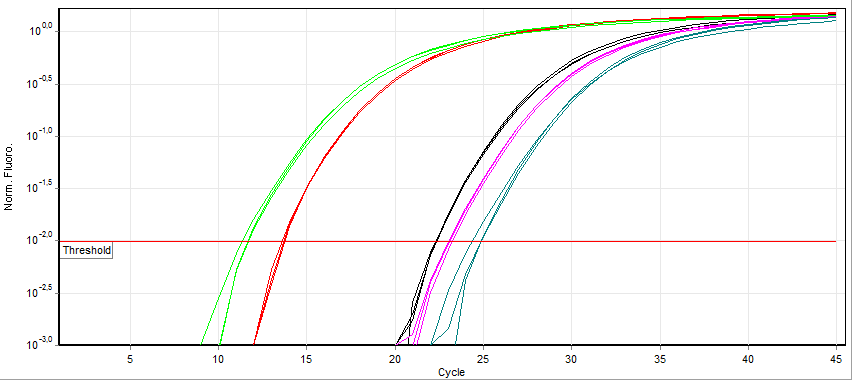
Real-time PCR
A real-time polymerase chain reaction (real-time PCR, or qPCR) is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR (i.e., in real time), not at its end, as in conventional PCR. Real-time PCR can be used quantitatively (quantitative real-time PCR) and semi-quantitatively (i.e., above/below a certain amount of DNA molecules) (semi-quantitative real-time PCR). Two common methods for the detection of PCR products in real-time PCR are (1) non-specific fluorescent dyes that intercalate with any double-stranded DNA and (2) sequence-specific DNA probes consisting of oligonucleotides that are labelled with a fluorescent reporter, which permits detection only after hybridization of the probe with its complementary sequence. The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines propose that the abbreviation ''qPCR'' be used for qu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microarray
A microarray is a multiplex lab-on-a-chip. Its purpose is to simultaneously detect the expression of thousands of genes from a sample (e.g. from a tissue). It is a two-dimensional array on a solid substrate—usually a glass slide or silicon thin-film cell—that assays (tests) large amounts of biological material using high-throughput screening miniaturized, multiplexed and parallel processing and detection methods. The concept and methodology of microarrays was first introduced and illustrated in antibody microarrays (also referred to as antibody matrix) by Tse Wen Chang in 1983 in a scientific publication and a series of patents. The " gene chip" industry started to grow significantly after the 1995 '' Science Magazine'' article by the Ron Davis and Pat Brown labs at Stanford University. With the establishment of companies, such as Affymetrix, Agilent, Applied Microarrays, Arrayjet, Illumina, and others, the technology of DNA microarrays has become the most sophistic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biotinylation
In biochemistry, biotinylation is the process of covalently attaching biotin to a protein, nucleic acid or other molecule. Biotinylation is rapid, specific and is unlikely to disturb the natural function of the molecule due to the small size of biotin (MW = 244.31 g/mol). Biotin binds to streptavidin and avidin with an extremely high affinity, fast on-rate, and high specificity, and these interactions are exploited in many areas of biotechnology to isolate biotinylated molecules of interest. Biotin-binding to streptavidin and avidin is resistant to extremes of heat, pH and proteolysis, making capture of biotinylated molecules possible in a wide variety of environments. Also, multiple biotin molecules can be conjugated to a protein of interest, which allows binding of multiple streptavidin, avidin or neutravidin protein molecules and increases the sensitivity of detection of the protein of interest. There is a large number of biotinylation reagents available that exploit t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Paramagnetism
Paramagnetism is a form of magnetism whereby some materials are weakly attracted by an externally applied magnetic field, and form internal, induced magnetic fields in the direction of the applied magnetic field. In contrast with this behavior, diamagnetic materials are repelled by magnetic fields and form induced magnetic fields in the direction opposite to that of the applied magnetic field. Paramagnetic materials include most chemical elements and some compounds; they have a relative magnetic permeability slightly greater than 1 (i.e., a small positive magnetic susceptibility) and hence are attracted to magnetic fields. The magnetic moment induced by the applied field is linear in the field strength and rather weak. It typically requires a sensitive analytical balance to detect the effect and modern measurements on paramagnetic materials are often conducted with a SQUID magnetometer. Paramagnetism is due to the presence of unpaired electrons in the material, so most atom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Streptavidin
Streptavidin is a 66.0 (tetramer) kDa protein purified from the bacterium '' Streptomyces avidinii''. Streptavidin homo-tetramers have an extraordinarily high affinity for biotin (also known as vitamin B7 or vitamin H). With a dissociation constant (Kd) on the order of ≈10−14 mol/L, the binding of biotin to streptavidin is one of the strongest non-covalent interactions known in nature. Streptavidin is used extensively in molecular biology and bionanotechnology due to the streptavidin-biotin complex's resistance to organic solvents, denaturants (e.g. guanidinium chloride), detergents (e.g. SDS, Triton X-100), proteolytic enzymes, and extremes of temperature and pH. Structure The crystal structure of streptavidin with biotin bound was reported by two groups in 1989. The structure was solved using multi wavelength anomalous diffraction by Hendrickson et al. at Columbia University and using multiple isomorphous replacement by Weber et al. at E. I. DuPont Central Resea ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Complementary Strand
A complement is something that completes something else. Complement may refer specifically to: The arts * Complement (music), an interval that, when added to another, spans an octave ** Aggregate complementation, the separation of pitch-class collections into complementary sets * Complementary color, in the visual arts Biology and medicine * Complement system (immunology), a cascade of proteins in the blood that form part of innate immunity *Complementary DNA, DNA reverse transcribed from a mature mRNA template *Complementarity (molecular biology), a property whereby double stranded nucleic acids pair with each other *Complementation (genetics), a test to determine if independent recessive mutant phenotypes are caused by mutations in the same gene or in different genes Grammar and linguistics * Complement (linguistics), a word or phrase having a particular syntactic role ** Subject complement, a word or phrase adding to a clause's subject after a linking verb * Phonetic c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Denaturation (biochemistry)
In biochemistry, denaturation is a process in which proteins or nucleic acids lose the quaternary structure, tertiary structure, and secondary structure which is present in their native state, by application of some external stress or compound such as a strong acid or base, a concentrated inorganic salt, an organic solvent (e.g., alcohol or chloroform), agitation and radiation or heat. If proteins in a living cell are denatured, this results in disruption of cell activity and possibly cell death. Protein denaturation is also a consequence of cell death. Denatured proteins can exhibit a wide range of characteristics, from conformational change and loss of solubility to aggregation due to the exposure of hydrophobic groups. The loss of solubility as a result of denaturation is called ''coagulation''. Denatured proteins lose their 3D structure and therefore cannot function. Protein folding is key to whether a globular or membrane protein can do its job correctly; it mu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Illumina (company)
Illumina, Inc. is an American biotechnology company, headquartered in San Diego, California. Incorporated on April 1, 1998, Illumina develops, manufactures, and markets integrated systems for the analysis of genetic variation and biological function. The company provides a line of products and services that serves the sequencing, genotyping and gene expression, and proteomics markets. Illumina's technology had purportedly reduced the cost of sequencing a human genome to by 2014. Its customers include genomic research centers, pharmaceutical companies, academic institutions, clinical research organizations, and biotechnology companies. History Illumina was founded in April 1998 by David Walt, Larry Bock, John Stuelpnagel, Anthony Czarnik, and Mark Chee. While working with CW Group, a venture-capital firm, Bock and Stuelpnagel uncovered what would become Illumina's BeadArray technology at Tufts University and negotiated an exclusive license to that technology. In 1999, Illumin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |







.jpg)