|
Proteogenomics Image 2
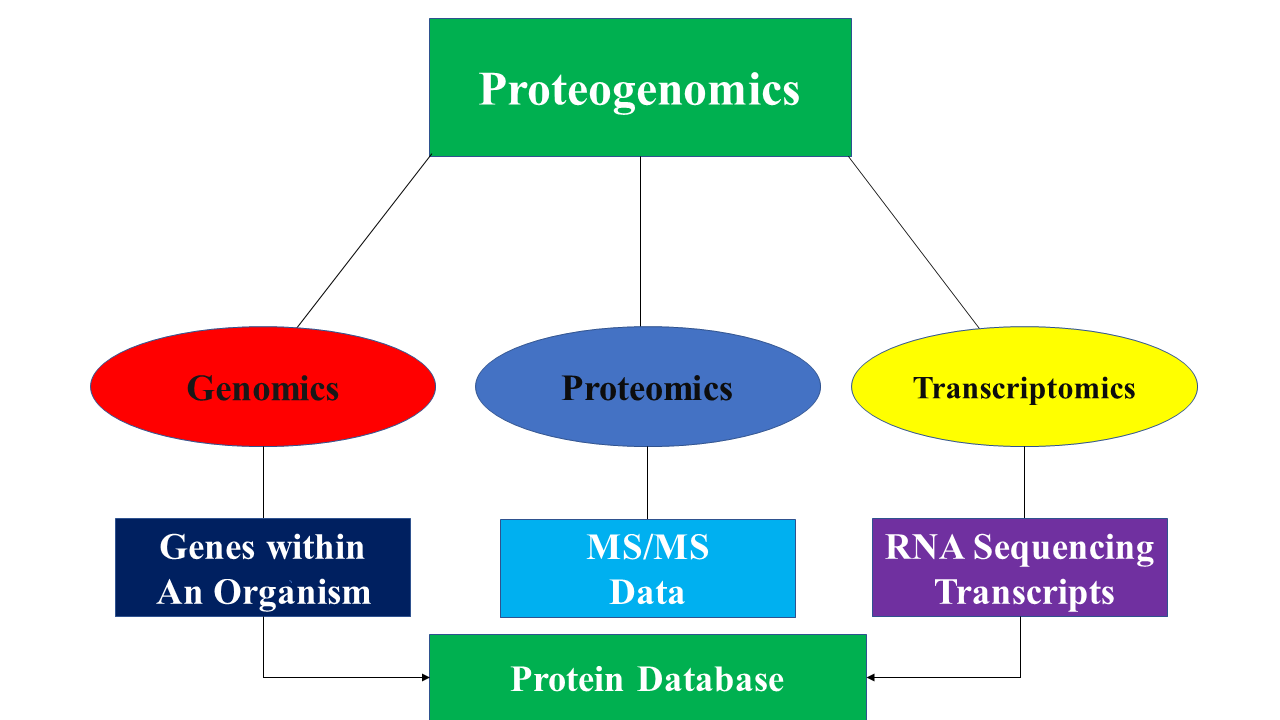
Proteogenomics is a field of biological research that utilizes a combination of proteomics, genomics, and transcriptomics to aid in the discovery and identification of Peptide, peptides. Proteogenomics is used to identify new peptides by comparing MS/MS spectra against a protein database that has been derived from genomic and transcriptomic information. Proteogenomics often refers to studies that use proteomic information, often derived from mass spectrometry, to improve gene annotations. The utilization of both proteomics and genomics data alongside advances in the availability and power of spectrographic and chromatographic technology led to the emergence of proteogenomics as its own field in 2004. Proteomics deals with proteins in the same way that Genomics studies the genetic code of entire organisms, while Transcriptomics technologies, Transcriptomics deals with the study of RNA sequencing and transcripts. While all three fields might use forms of mass spectrometry and chromato ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteogenomics Image 2
Proteogenomics is a field of biological research that utilizes a combination of proteomics, genomics, and transcriptomics to aid in the discovery and identification of Peptide, peptides. Proteogenomics is used to identify new peptides by comparing MS/MS spectra against a protein database that has been derived from genomic and transcriptomic information. Proteogenomics often refers to studies that use proteomic information, often derived from mass spectrometry, to improve gene annotations. The utilization of both proteomics and genomics data alongside advances in the availability and power of spectrographic and chromatographic technology led to the emergence of proteogenomics as its own field in 2004. Proteomics deals with proteins in the same way that Genomics studies the genetic code of entire organisms, while Transcriptomics technologies, Transcriptomics deals with the study of RNA sequencing and transcripts. While all three fields might use forms of mass spectrometry and chromato ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Open Reading Frame
In molecular biology, open reading frames (ORFs) are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a prokaryotic DNA sequence, where only one of the six possible reading frames will be "open" (the "reading", however, refers to the RNA produced by transcription of the DNA and its subsequent interaction with the ribosome in translation). Such an ORF may contain a start codon (usually AUG in terms of RNA) and by definition cannot extend beyond a stop codon (usually UAA, UAG or UGA in RNA). That start codon (not necessarily the first) indicates where translation may start. The transcription termination site is located after the ORF, beyond the translation stop codon. If transcription were to cease before the stop codon, an incomplete protein would be made during translation. In eukaryotic genes with multiple exons, introns are removed and exons are then joined together after transcription to yield the final ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Post-translational Modification
Post-translational modification (PTM) is the covalent and generally enzymatic modification of proteins following protein biosynthesis. This process occurs in the endoplasmic reticulum and the golgi apparatus. Proteins are synthesized by ribosomes translating mRNA into polypeptide chains, which may then undergo PTM to form the mature protein product. PTMs are important components in cell signaling, as for example when prohormones are converted to hormones. Post-translational modifications can occur on the amino acid side chains or at the protein's C- or N- termini. They can extend the chemical repertoire of the 20 standard amino acids by modifying an existing functional group or introducing a new one such as phosphate. Phosphorylation is a highly effective mechanism for regulating the activity of enzymes and is the most common post-translational modification. Many eukaryotic and prokaryotic proteins also have carbohydrate molecules attached to them in a process called glycosyla ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteolysis
Proteolysis is the breakdown of proteins into smaller polypeptides or amino acids. Uncatalysed, the hydrolysis of peptide bonds is extremely slow, taking hundreds of years. Proteolysis is typically catalysed by cellular enzymes called proteases, but may also occur by intra-molecular digestion. Proteolysis in organisms serves many purposes; for example, digestive enzymes break down proteins in food to provide amino acids for the organism, while proteolytic processing of a polypeptide chain after its synthesis may be necessary for the production of an active protein. It is also important in the regulation of some physiological and cellular processes including apoptosis, as well as preventing the accumulation of unwanted or misfolded proteins in cells. Consequently, abnormality in the regulation of proteolysis can cause disease. Proteolysis can also be used as an analytical tool for studying proteins in the laboratory, and it may also be used in industry, for example in food proc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Signal Peptide
A signal peptide (sometimes referred to as signal sequence, targeting signal, localization signal, localization sequence, transit peptide, leader sequence or leader peptide) is a short peptide (usually 16-30 amino acids long) present at the N-terminus (or occasionally nonclassically at the C-terminus or internally) of most newly synthesized proteins that are destined toward the secretory pathway. These proteins include those that reside either inside certain organelles (the endoplasmic reticulum, Golgi or endosomes), secreted from the cell, or inserted into most cellular membranes. Although most type I membrane-bound proteins have signal peptides, the majority of type II and multi-spanning membrane-bound proteins are targeted to the secretory pathway by their first transmembrane domain, which biochemically resembles a signal sequence except that it is not cleaved. They are a kind of target peptide. Function (translocation) Signal peptides function to prompt a cell to translo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methionine
Methionine (symbol Met or M) () is an essential amino acid in humans. As the precursor of other amino acids such as cysteine and taurine, versatile compounds such as SAM-e, and the important antioxidant glutathione, methionine plays a critical role in the metabolism and health of many species, including humans. It is encoded by the codon AUG. Methionine is also an important part of angiogenesis, the growth of new blood vessels. Supplementation may benefit those suffering from copper poisoning. Overconsumption of methionine, the methyl group donor in DNA methylation, is related to cancer growth in a number of studies. Methionine was first isolated in 1921 by John Howard Mueller. Biochemical details Methionine (abbreviated as Met or M; encoded by the codon AUG) is an α-amino acid that is used in the biosynthesis of proteins. It contains a carboxyl group (which is in the deprotonated −COO− form under biological pH conditions), an amino group (which is in the protonated fo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Frameshift
Ribosomal frameshifting, also known as translational frameshifting or translational recoding, is a biological phenomenon that occurs during translation that results in the production of multiple, unique proteins from a single mRNA. The process can be programmed by the nucleotide sequence of the mRNA and is sometimes affected by the secondary, 3-dimensional mRNA structure. It has been described mainly in viruses (especially retroviruses), retrotransposons and bacterial insertion elements, and also in some cellular genes. Process overview Proteins are translated by reading tri-nucleotides on the mRNA strand, also known as codons, from one end of the mRNA to the other (from the 5' to the 3' end) starting with the amino acid methionine as the start (initiation) codon AUG. Each codon is translated into a single amino acid. The code itself is considered degenerate, meaning that a particular amino acid can be specified by more than one codons. However, a shift of any number of nucleo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Shewanella
''Shewanella'' is the sole genus included in the marine bacteria family Shewanellaceae. Some species within it were formerly classed as ''Alteromonas''. ''Shewanella'' consists of facultatively anaerobic Gram-negative rods, most of which are found in extreme aquatic habitats where the temperature is very low and the pressure is very high. ''Shewanella'' bacteria are a normal component of the surface flora of fish and are implicated in fish spoilage. ''Shewanella chilikensis'', a species of the genus ''Shewanella'' commonly found in the marine sponges of Saint Martin's Island of the Bay of Bengal, Bangladesh. ''Shewanella oneidensis'' MR-1 is a widely used laboratory model to study anaerobic respiration of metals and other anaerobic extracellular electron acceptors, and for teaching about microbial electrogenesis and microbial fuel cells. Biochemical characteristics of ''Shewanella'' species Colony, morphological, physiological, and biochemical characteristics of ''Shewanell ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mycobacterium
''Mycobacterium'' is a genus of over 190 species in the phylum Actinomycetota, assigned its own family, Mycobacteriaceae. This genus includes pathogens known to cause serious diseases in mammals, including tuberculosis ('' M. tuberculosis'') and leprosy ('' M. leprae'') in humans. The Greek prefix ''myco-'' means 'fungus', alluding to this genus' mold-like colony surfaces. Since this genus has cell walls with Gram-positive and Gram-negative features, acid-fast staining is used to emphasize their resistance to acids, compared to other cell types. Metabolism and Morphology Mycobacteria are aerobic with 0.2-0.6 µm wide and 1.0-10 µm long rod shapes. They are generally non-motile, except for the species ''Mycobacterium marinum'', which has been shown to be motile within macrophages. Mycobacteria possess capsules and most do not form endospores. ''M. marinum'' and perhaps ''M. bovis'' have been shown to sporulate; however, this has been contested by further research. The disti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Escherichia Coli
''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus ''Escherichia'' that is commonly found in the lower intestine of warm-blooded organisms. Most ''E. coli'' strains are harmless, but some serotypes ( EPEC, ETEC etc.) can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. Most strains do not cause disease in humans and are part of the normal microbiota of the gut; such strains are harmless or even beneficial to humans (although these strains tend to be less studied than the pathogenic ones). For example, some strains of ''E. coli'' benefit their hosts by producing vitamin K2 or by preventing the colonization of the intestine by pathogenic bacteria. These mutually beneficial relationships between ''E. col ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Expressed Sequence Tag
In genetics, an expressed sequence tag (EST) is a short sub-sequence of a cDNA sequence. ESTs may be used to identify gene transcripts, and were instrumental in gene discovery and in gene-sequence determination. The identification of ESTs has proceeded rapidly, with approximately 74.2 million ESTs now available in public databases (e.g. GenBank 1 January 2013, all species). EST approaches have largely been superseded by whole genome and transcriptome sequencing and metagenome sequencing. An EST results from one-shot sequencing of a cloned cDNA. The cDNAs used for EST generation are typically individual clones from a cDNA library. The resulting sequence is a relatively low-quality fragment whose length is limited by current technology to approximately 500 to 800 nucleotides. Because these clones consist of DNA that is complementary to mRNA, the ESTs represent portions of expressed genes. They may be represented in databases as either cDNA/mRNA sequence or as the reverse complement of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Coding Regions
The coding region of a gene, also known as the coding sequence (CDS), is the portion of a gene's DNA or RNA that codes for protein. Studying the length, composition, regulation, splicing, structures, and functions of coding regions compared to non-coding regions over different species and time periods can provide a significant amount of important information regarding gene organization and evolution of prokaryotes and eukaryotes. This can further assist in mapping the Human Genome Project, human genome and developing gene therapy. Definition Although this term is also sometimes used interchangeably with exon, it is not the exact same thing: the exon is composed of the coding region as well as the 3' and 5' untranslated regions of the RNA, and so therefore, an exon would be partially made up of coding regions. The 3' and 5' untranslated regions of the RNA, which do not code for protein, are termed Non-coding region, non-coding regions and are not discussed on this page. There is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |