Post-translational modification on:
[Wikipedia]
[Google]
[Amazon]
 In
In
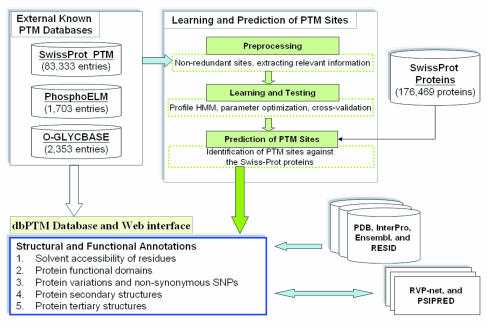 Protein sequences contain sequence motifs that are recognized by modifying enzymes, and which can be documented or predicted in PTM databases. With the large number of different modifications being discovered, there is a need to document this sort of information in databases. PTM information can be collected through experimental means or predicted from high-quality, manually curated data. Numerous databases have been created, often with a focus on certain taxonomic groups (e.g. human proteins) or other features.
Protein sequences contain sequence motifs that are recognized by modifying enzymes, and which can be documented or predicted in PTM databases. With the large number of different modifications being discovered, there is a need to document this sort of information in databases. PTM information can be collected through experimental means or predicted from high-quality, manually curated data. Numerous databases have been created, often with a focus on certain taxonomic groups (e.g. human proteins) or other features.
PhosphoSitePlus
– A database of comprehensive information and tools for the study of mammalian protein post-translational modification * ProteomeScout – A database of proteins and post-translational modifications experimentally * Human Protein Reference Database – A database for different modifications and understand different proteins, their class, and function/process related to disease causing proteins * PROSITE – A database of Consensus patterns for many types of PTM's including sites * Protein Information Resource, RESID – A database consisting of a collection of annotations and structures for PTMs.
iPTMnet
– A database that integrates PTM information from several knowledgbases and text mining results. * dbPTM – A database that shows different PTM's and information regarding their chemical components/structures and a frequency for amino acid modified site
Uniprot
has PTM information although that may be less comprehensive than in more specialized databases.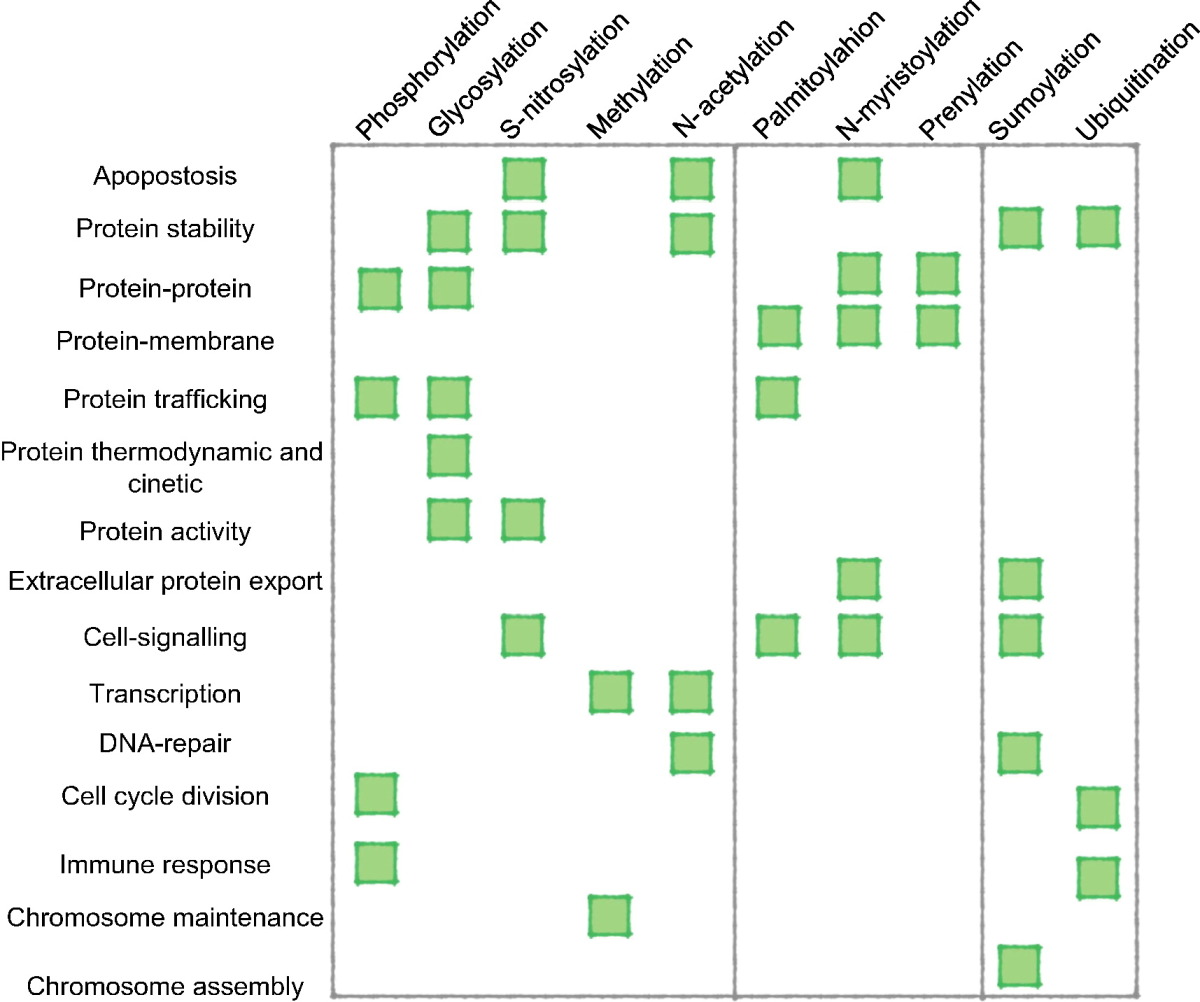
The ''O''-GlcNAc Database
- A curated database for protein O-GlcNAcylation and referencing more than 14 000 protein entries and 10 000 ''O''-GlcNAc sites.
Controlled vocabulary of post-translational modifications
in Uniprot
List of posttranslational modifications in ExPASy
Browse SCOP domains by PTM
— from the dcGO database
Overview and description of commonly used post-translational modification detection techniques
{{DEFAULTSORT:Posttranslational Modification Gene expression Protein structure Protein biosynthesis Post-translational modification, Cell biology
molecular biology
Molecular biology is a branch of biology that seeks to understand the molecule, molecular basis of biological activity in and between Cell (biology), cells, including biomolecule, biomolecular synthesis, modification, mechanisms, and interactio ...
, post-translational modification (PTM) is the covalent
A covalent bond is a chemical bond that involves the sharing of electrons to form electron pairs between atoms. These electron pairs are known as shared pairs or bonding pairs. The stable balance of attractive and repulsive forces between atom ...
process of changing protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
s following protein biosynthesis
Protein biosynthesis, or protein synthesis, is a core biological process, occurring inside Cell (biology), cells, homeostasis, balancing the loss of cellular proteins (via Proteolysis, degradation or Protein targeting, export) through the produc ...
. PTMs may involve enzymes
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as pro ...
or occur spontaneously. Proteins are created by ribosomes
Ribosomes () are macromolecular machines, found within all cells, that perform biological protein synthesis (messenger RNA translation). Ribosomes link amino acids together in the order specified by the codons of messenger RNA molecules to fo ...
, which translate mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein.
mRNA is ...
into polypeptide chain
Peptides are short chains of amino acids linked by peptide bonds. A polypeptide is a longer, continuous, unbranched peptide chain. Polypeptides that have a molecular mass of 10,000 Da or more are called proteins. Chains of fewer than twenty ami ...
s, which may then change to form the mature protein product. PTMs are important components in cell signalling
A signal is both the process and the result of transmission of data over some media accomplished by embedding some variation. Signals are important in multiple subject fields including signal processing, information theory and biology.
In ...
, as for example when prohormone
A prohormone is a committed precursor of a hormone consisting of peptide hormones synthesized together that has a minimal hormonal effect by itself because of its expression-suppressing structure, often created by protein folding and binding addit ...
s are converted to hormone
A hormone (from the Ancient Greek, Greek participle , "setting in motion") is a class of cell signaling, signaling molecules in multicellular organisms that are sent to distant organs or tissues by complex biological processes to regulate physio ...
s.
Post-translational modifications can occur on the amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
side chain
In organic chemistry and biochemistry, a side chain is a substituent, chemical group that is attached to a core part of the molecule called the "main chain" or backbone chain, backbone. The side chain is a hydrocarbon branching element of a mo ...
s or at the protein's C- or N- termini. They can expand the chemical set of the 22 amino acids
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the Proteinogenic amino acid, 22 α-amino acids incorporated into p ...
by changing an existing functional group
In organic chemistry, a functional group is any substituent or moiety (chemistry), moiety in a molecule that causes the molecule's characteristic chemical reactions. The same functional group will undergo the same or similar chemical reactions r ...
or adding a new one such as phosphate. Phosphorylation
In biochemistry, phosphorylation is described as the "transfer of a phosphate group" from a donor to an acceptor. A common phosphorylating agent (phosphate donor) is ATP and a common family of acceptor are alcohols:
:
This equation can be writ ...
is highly effective for controlling the enzyme activity and is the most common change after translation. Many eukaryotic
The eukaryotes ( ) constitute the Domain (biology), domain of Eukaryota or Eukarya, organisms whose Cell (biology), cells have a membrane-bound cell nucleus, nucleus. All animals, plants, Fungus, fungi, seaweeds, and many unicellular organisms ...
and prokaryotic
A prokaryote (; less commonly spelled procaryote) is a single-celled organism whose cell lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Ancient Greek (), meaning 'before', and (), meaning 'nut' ...
proteins also have carbohydrate
A carbohydrate () is a biomolecule composed of carbon (C), hydrogen (H), and oxygen (O) atoms. The typical hydrogen-to-oxygen atomic ratio is 2:1, analogous to that of water, and is represented by the empirical formula (where ''m'' and ''n'' ...
molecules attached to them in a process called glycosylation
Glycosylation is the reaction in which a carbohydrate (or ' glycan'), i.e. a glycosyl donor, is attached to a hydroxyl or other functional group of another molecule (a glycosyl acceptor) in order to form a glycoconjugate. In biology (but not ...
, which can promote protein folding
Protein folding is the physical process by which a protein, after Protein biosynthesis, synthesis by a ribosome as a linear chain of Amino acid, amino acids, changes from an unstable random coil into a more ordered protein tertiary structure, t ...
and improve stability as well as serving regulatory functions. Attachment of lipid
Lipids are a broad group of organic compounds which include fats, waxes, sterols, fat-soluble vitamins (such as vitamins A, D, E and K), monoglycerides, diglycerides, phospholipids, and others. The functions of lipids include storing ...
molecules, known as lipidation, often targets a protein or part of a protein attached to the cell membrane
The cell membrane (also known as the plasma membrane or cytoplasmic membrane, and historically referred to as the plasmalemma) is a biological membrane that separates and protects the interior of a cell from the outside environment (the extr ...
.
Other forms of post-translational modification consist of cleaving peptide bond
In organic chemistry, a peptide bond is an amide type of covalent chemical bond linking two consecutive alpha-amino acids from C1 (carbon number one) of one alpha-amino acid and N2 (nitrogen number two) of another, along a peptide or protein cha ...
s, as in processing a propeptide
A protein precursor, also called a pro-protein or pro-peptide, is an inactive protein (or peptide) that can be turned into an active form by post-translational modification, such as breaking off a piece of the molecule or adding on another molecule ...
to a mature form or removing the initiator methionine
Methionine (symbol Met or M) () is an essential amino acid in humans.
As the precursor of other non-essential amino acids such as cysteine and taurine, versatile compounds such as SAM-e, and the important antioxidant glutathione, methionine play ...
residue. The formation of disulfide bond
In chemistry, a disulfide (or disulphide in British English) is a compound containing a functional group or the anion. The linkage is also called an SS-bond or sometimes a disulfide bridge and usually derived from two thiol groups.
In inor ...
s from cysteine
Cysteine (; symbol Cys or C) is a semiessential proteinogenic amino acid with the chemical formula, formula . The thiol side chain in cysteine enables the formation of Disulfide, disulfide bonds, and often participates in enzymatic reactions as ...
residues may also be referred to as a post-translational modification. For instance, the peptide hormone
A hormone (from the Ancient Greek, Greek participle , "setting in motion") is a class of cell signaling, signaling molecules in multicellular organisms that are sent to distant organs or tissues by complex biological processes to regulate physio ...
insulin
Insulin (, from Latin ''insula'', 'island') is a peptide hormone produced by beta cells of the pancreatic islets encoded in humans by the insulin (''INS)'' gene. It is the main Anabolism, anabolic hormone of the body. It regulates the metabol ...
is cut twice after disulfide bonds are formed, and a propeptide
A protein precursor, also called a pro-protein or pro-peptide, is an inactive protein (or peptide) that can be turned into an active form by post-translational modification, such as breaking off a piece of the molecule or adding on another molecule ...
is removed from the middle of the chain; the resulting protein consists of two polypeptide chains connected by disulfide bonds.
Some types of post-translational modification are consequences of oxidative stress
Oxidative stress reflects an imbalance between the systemic manifestation of reactive oxygen species and a biological system's ability to readily detoxify the reactive intermediates or to repair the resulting damage. Disturbances in the normal ...
. Carbonylation
In chemistry, carbonylation refers to reactions that introduce carbon monoxide (CO) into organic and inorganic substrates. Carbon monoxide is abundantly available and conveniently reactive, so it is widely used as a reactant in industrial chemis ...
is one example that targets the modified protein for degradation and can result in the formation of protein aggregates. Specific amino acid modifications can be used as biomarker
In biomedical contexts, a biomarker, or biological marker, is a measurable indicator of some biological state or condition. Biomarkers are often measured and evaluated using blood, urine, or soft tissues to examine normal biological processes, ...
s indicating oxidative damage.
PTMs and metal ions play a crucial and reciprocal role in regulating protein function, influencing cellular processes such as signal transduction and gene expression, with dysregulated interactions implicated in diseases like cancer and neurodegenerative disorders.
Sites that often undergo post-translational modification are those that have a functional group that can serve as a nucleophile
In chemistry, a nucleophile is a chemical species that forms bonds by donating an electron pair. All molecules and ions with a free pair of electrons or at least one pi bond can act as nucleophiles. Because nucleophiles donate electrons, they are ...
in the reaction: the hydroxyl
In chemistry, a hydroxy or hydroxyl group is a functional group with the chemical formula and composed of one oxygen atom covalently bonded to one hydrogen atom. In organic chemistry, alcohols and carboxylic acids contain one or more hydroxy ...
groups of serine
Serine
(symbol Ser or S) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α- amino group (which is in the protonated − form under biological conditions), a carboxyl group (which is in the deprotonated − ...
, threonine
Threonine (symbol Thr or T) is an amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form when dissolved in water), a carboxyl group (which is in the deprotonated −COO− ...
, and tyrosine
-Tyrosine or tyrosine (symbol Tyr or Y) or 4-hydroxyphenylalanine is one of the 20 standard amino acids that are used by cells to synthesize proteins. It is a conditionally essential amino acid with a polar side group. The word "tyrosine" is ...
; the amine
In chemistry, amines (, ) are organic compounds that contain carbon-nitrogen bonds. Amines are formed when one or more hydrogen atoms in ammonia are replaced by alkyl or aryl groups. The nitrogen atom in an amine possesses a lone pair of elec ...
forms of lysine
Lysine (symbol Lys or K) is an α-amino acid that is a precursor to many proteins. Lysine contains an α-amino group (which is in the protonated form when the lysine is dissolved in water at physiological pH), an α-carboxylic acid group ( ...
, arginine
Arginine is the amino acid with the formula (H2N)(HN)CN(H)(CH2)3CH(NH2)CO2H. The molecule features a guanidinium, guanidino group appended to a standard amino acid framework. At physiological pH, the carboxylic acid is deprotonated (−CO2−) a ...
, and histidine
Histidine (symbol His or H) is an essential amino acid that is used in the biosynthesis of proteins. It contains an Amine, α-amino group (which is in the protonated –NH3+ form under Physiological condition, biological conditions), a carboxylic ...
; the thiolate
In organic chemistry, a thiol (; ), or thiol derivative, is any organosulfur compound of the form , where R represents an alkyl or other organic substituent. The functional group itself is referred to as either a thiol group or a sulfhydryl grou ...
anion
An ion () is an atom or molecule with a net electrical charge. The charge of an electron is considered to be negative by convention and this charge is equal and opposite to the charge of a proton, which is considered to be positive by conven ...
of cysteine
Cysteine (; symbol Cys or C) is a semiessential proteinogenic amino acid with the chemical formula, formula . The thiol side chain in cysteine enables the formation of Disulfide, disulfide bonds, and often participates in enzymatic reactions as ...
; the carboxylate
In organic chemistry, a carboxylate is the conjugate base of a carboxylic acid, (or ). It is an anion, an ion with negative charge.
Carboxylate salts are salts that have the general formula , where M is a metal and ''n'' is 1, 2,... ...
s of aspartate
Aspartic acid (symbol Asp or D; the ionic form is known as aspartate), is an α-amino acid that is used in the biosynthesis of proteins. The L-isomer of aspartic acid is one of the 22 proteinogenic amino acids, i.e., the building blocks of protein ...
and glutamate
Glutamic acid (symbol Glu or E; known as glutamate in its anionic form) is an α-amino acid that is used by almost all living beings in the biosynthesis of proteins. It is a Essential amino acid, non-essential nutrient for humans, meaning that ...
; and the N- and C-termini. In addition, although the amide
In organic chemistry, an amide, also known as an organic amide or a carboxamide, is a chemical compound, compound with the general formula , where R, R', and R″ represent any group, typically organyl functional group, groups or hydrogen at ...
of asparagine
Asparagine (symbol Asn or N) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form under biological conditions), an α-carboxylic acid group (which is in the depro ...
is a weak nucleophile, it can serve as an attachment point for glycan
The terms glycans and polysaccharides are defined by IUPAC as synonyms meaning "compounds consisting of a large number of monosaccharides linked glycosidically". However, in practice the term glycan may also be used to refer to the carbohydrate ...
s. Rarer modifications can occur at oxidized methionine
Methionine (symbol Met or M) () is an essential amino acid in humans.
As the precursor of other non-essential amino acids such as cysteine and taurine, versatile compounds such as SAM-e, and the important antioxidant glutathione, methionine play ...
s and at some methylene groups in side chains.
Post-translational modification of proteins can be experimentally detected by a variety of techniques, including mass spectrometry
Mass spectrometry (MS) is an analytical technique that is used to measure the mass-to-charge ratio of ions. The results are presented as a ''mass spectrum'', a plot of intensity as a function of the mass-to-charge ratio. Mass spectrometry is used ...
, Eastern blotting
The eastern blot, or eastern blotting, is a biochemical technique used to analyze protein post-translational modifications including the addition of lipids, phosphates, and glycoconjugates. It is most often used to detect carbohydrate epitopes. Thu ...
, and Western blotting
The western blot (sometimes called the protein immunoblot), or western blotting, is a widely used analytical technique in molecular biology and immunogenetics to detect specific proteins in a sample of tissue homogenate or extract. Besides detec ...
.
PTMs involving addition of functional groups
Addition by an enzyme ''in vivo''
Hydrophobic groups for membrane localization
* myristoylation (a type ofacylation
In chemistry, acylation is a broad class of chemical reactions in which an acyl group () is added to a substrate. The compound providing the acyl group is called the acylating agent. The substrate to be acylated and the product include the foll ...
), attachment of myristate, a C14 saturated acid
* palmitoylation (a type of acylation), attachment of palmitate
Palmitic acid (hexadecanoic acid in IUPAC nomenclature) is a fatty acid with a 16-carbon chain. It is the most common saturated fatty acid found in animals, plants and microorganisms.Gunstone, F. D., John L. Harwood, and Albert J. Dijkstra. The Li ...
, a C16 saturated acid
* isoprenylation
Prenylation (also known as isoprenylation or lipidation) is the addition of hydrophobic molecules to a protein or a biomolecule. It is usually assumed that prenyl groups (3-methylbut-2-en-1-yl) facilitate attachment to cell membranes, similar to ...
or prenylation
Prenylation (also known as isoprenylation or lipidation) is the addition of hydrophobic molecules to a protein or a biomolecule. It is usually assumed that prenyl groups (3-methylbut-2-en-1-yl) facilitate attachment to cell membranes, similar to ...
, the addition of an isoprenoid group (e.g. farnesol and geranylgeraniol
Geranylgeraniol is a diterpenoid alcohol. It is a colorless waxy solid. It is an important intermediate in the biosynthesis of other diterpenes, of vitamins E, and of K. It is a derivative of geranylgeraniol pyrophosphate, which is a precursor ...
)
** farnesylation
** geranylgeranylation Geranylgeranylation is a form of prenylation, which is a post-translational modification of proteins that involves the attachment of one or two 20-carbon lipophilic geranylgeranyl isoprenoid units from geranylgeranyl diphosphate to one or two cys ...
* glypiation, glycosylphosphatidylinositol (GPI) anchor formation via an amide bond to C-terminal tail
Cofactors for enhanced enzymatic activity
* lipoylation (a type of acylation), attachment of alipoate
Lipoic acid (LA), also known as α-lipoic acid, alpha-lipoic acid (ALA) and thioctic acid, is an organosulfur compound derived from caprylic acid (octanoic acid). ALA, which is made in animals normally, is essential for aerobic metabolism. It i ...
(C8) functional group
* flavin moiety (flavin mononucleotide
Flavin mononucleotide (FMN), or riboflavin-5′-phosphate, is a biomolecule produced from riboflavin (vitamin B2) by the enzyme riboflavin kinase and functions as the prosthetic group of various oxidoreductases, including NADH dehydrogenase, as ...
(FMN) or flavin adenine dinucleotide
In biochemistry, flavin adenine dinucleotide (FAD) is a redox-active coenzyme associated with various proteins, which is involved with several enzymatic reactions in metabolism. A flavoprotein is a protein that contains a flavin group, which ma ...
(FAD)) may be covalently attached
* heme C attachment via thioether
In organic chemistry, a sulfide (British English sulphide) or thioether is an organosulfur functional group with the connectivity as shown on right. Like many other sulfur-containing compounds, Volatile organic compound, volatile sulfides have ...
bonds with cysteine
Cysteine (; symbol Cys or C) is a semiessential proteinogenic amino acid with the chemical formula, formula . The thiol side chain in cysteine enables the formation of Disulfide, disulfide bonds, and often participates in enzymatic reactions as ...
s
* phosphopantetheinylation, the addition of a 4'-phosphopantetheinyl moiety from coenzyme A
Coenzyme A (CoA, SHCoA, CoASH) is a coenzyme, notable for its role in the Fatty acid metabolism#Synthesis, synthesis and Fatty acid metabolism#.CE.B2-Oxidation, oxidation of fatty acids, and the oxidation of pyruvic acid, pyruvate in the citric ac ...
, as in fatty acid, polyketide, non-ribosomal peptide and leucine biosynthesis
* retinylidene
Retinal (also known as retinaldehyde) is a polyene chromophore. Retinal, bound to proteins called opsins, is the chemical basis of visual phototransduction, the light-detection stage of visual perception (vision).
Some microorganisms use retin ...
Schiff base
In organic chemistry, a Schiff base (named after Hugo Schiff) is a compound with the general structure ( = alkyl or aryl, but not hydrogen). They can be considered a sub-class of imines, being either secondary ketimines or secondary aldim ...
formation
Modifications of translation factors
* diphthamide formation (on a histidine found in eEF2) * ethanolamine phosphoglycerol attachment (on glutamate found in eEF1α) * hypusine formation (on conserved lysine of eIF5A (eukaryotic) and aIF5A (archaeal)) * beta-Lysine addition on a conserved lysine of the elongation factor P (EFP) in most bacteria. EFP is a homolog to eIF5A (eukaryotic) and aIF5A (archaeal) (see above).Smaller chemical groups
*acylation
In chemistry, acylation is a broad class of chemical reactions in which an acyl group () is added to a substrate. The compound providing the acyl group is called the acylating agent. The substrate to be acylated and the product include the foll ...
, e.g. ''O''-acylation (esters
In chemistry, an ester is a chemical compound, compound derived from an acid (either organic or inorganic) in which the hydrogen atom (H) of at least one acidic hydroxyl group () of that acid is replaced by an organyl group (R). These compounds c ...
), ''N''-acylation (amides
In organic chemistry, an amide, also known as an organic amide or a carboxamide, is a compound with the general formula , where R, R', and R″ represent any group, typically organyl groups or hydrogen atoms. The amide group is called a p ...
), ''S''-acylation ( thioesters)
** acetylation
:
In chemistry, acetylation is an organic esterification reaction with acetic acid. It introduces an acetyl group into a chemical compound. Such compounds are termed ''acetate esters'' or simply ''acetates''. Deacetylation is the opposite react ...
, the addition of an acetyl
In organic chemistry, an acetyl group is a functional group denoted by the chemical formula and the structure . It is sometimes represented by the symbol Ac (not to be confused with the element actinium). In IUPAC nomenclature, an acetyl grou ...
group, either at the N-terminus
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amin ...
of the protein or at lysine
Lysine (symbol Lys or K) is an α-amino acid that is a precursor to many proteins. Lysine contains an α-amino group (which is in the protonated form when the lysine is dissolved in water at physiological pH), an α-carboxylic acid group ( ...
residues. The reverse is called deacetylation
:
In chemistry, acetylation is an organic esterification reaction with acetic acid. It introduces an acetyl group into a chemical compound. Such compounds are termed ''acetate esters'' or simply ''acetates''. Deacetylation is the opposite react ...
.
** formylation
* alkylation Alkylation is a chemical reaction that entails transfer of an alkyl group. The alkyl group may be transferred as an alkyl carbocation, a free radical, a carbanion, or a carbene (or their equivalents). Alkylating agents are reagents for effecting al ...
, the addition of an alkyl
In organic chemistry, an alkyl group is an alkane missing one hydrogen.
The term ''alkyl'' is intentionally unspecific to include many possible substitutions.
An acyclic alkyl has the general formula of . A cycloalkyl group is derived from a cy ...
group, e.g. methyl
In organic chemistry, a methyl group is an alkyl derived from methane, containing one carbon atom bonded to three hydrogen atoms, having chemical formula (whereas normal methane has the formula ). In formulas, the group is often abbreviated as ...
, ethyl
** methylation
Methylation, in the chemistry, chemical sciences, is the addition of a methyl group on a substrate (chemistry), substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replac ...
the addition of a methyl
In organic chemistry, a methyl group is an alkyl derived from methane, containing one carbon atom bonded to three hydrogen atoms, having chemical formula (whereas normal methane has the formula ). In formulas, the group is often abbreviated as ...
group, usually at lysine
Lysine (symbol Lys or K) is an α-amino acid that is a precursor to many proteins. Lysine contains an α-amino group (which is in the protonated form when the lysine is dissolved in water at physiological pH), an α-carboxylic acid group ( ...
or arginine
Arginine is the amino acid with the formula (H2N)(HN)CN(H)(CH2)3CH(NH2)CO2H. The molecule features a guanidinium, guanidino group appended to a standard amino acid framework. At physiological pH, the carboxylic acid is deprotonated (−CO2−) a ...
residues. The reverse is called demethylation
Demethylation is the chemical process resulting in the removal of a methyl group (CH3) from a molecule. A common way of demethylation is the replacement of a methyl group by a hydrogen atom, resulting in a net loss of one carbon and two hydrogen at ...
.
* amidation at C-terminus. Formed by oxidative dissociation of a C-terminal Gly residue.
* amide
In organic chemistry, an amide, also known as an organic amide or a carboxamide, is a chemical compound, compound with the general formula , where R, R', and R″ represent any group, typically organyl functional group, groups or hydrogen at ...
bond formation
** amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
addition
*** arginylation, a tRNA
Transfer ribonucleic acid (tRNA), formerly referred to as soluble ribonucleic acid (sRNA), is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes). In a cell, it provides the physical link between the gene ...
-mediation addition
*** polyglutamylation, covalent linkage of glutamic acid
Glutamic acid (symbol Glu or E; known as glutamate in its anionic form) is an α- amino acid that is used by almost all living beings in the biosynthesis of proteins. It is a non-essential nutrient for humans, meaning that the human body can ...
residues to the N-terminus of tubulin and some other proteins. (See tubulin polyglutamylase)
*** polyglycylation, covalent linkage of one to more than 40 glycine
Glycine (symbol Gly or G; ) is an amino acid that has a single hydrogen atom as its side chain. It is the simplest stable amino acid. Glycine is one of the proteinogenic amino acids. It is encoded by all the codons starting with GG (G ...
residues to the tubulin
Tubulin in molecular biology can refer either to the tubulin protein superfamily of globular proteins, or one of the member proteins of that superfamily. α- and β-tubulins polymerize into microtubules, a major component of the eukaryotic cytosk ...
C-terminal tail
* butyrylation
* gamma-carboxylation dependent on Vitamin K
Vitamin K is a family of structurally similar, fat-soluble vitamers found in foods and marketed as dietary supplements. The human body requires vitamin K for post-translational modification, post-synthesis modification of certain proteins ...
* glycosylation
Glycosylation is the reaction in which a carbohydrate (or ' glycan'), i.e. a glycosyl donor, is attached to a hydroxyl or other functional group of another molecule (a glycosyl acceptor) in order to form a glycoconjugate. In biology (but not ...
, the addition of a glycosyl
In organic chemistry, a glycosyl group is a univalent free radical or substituent structure obtained by removing the hydroxyl () group from the hemiacetal () group found in the cyclic form of a monosaccharide and, by extension, of a lower ol ...
group to either arginine
Arginine is the amino acid with the formula (H2N)(HN)CN(H)(CH2)3CH(NH2)CO2H. The molecule features a guanidinium, guanidino group appended to a standard amino acid framework. At physiological pH, the carboxylic acid is deprotonated (−CO2−) a ...
, asparagine
Asparagine (symbol Asn or N) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form under biological conditions), an α-carboxylic acid group (which is in the depro ...
, cysteine
Cysteine (; symbol Cys or C) is a semiessential proteinogenic amino acid with the chemical formula, formula . The thiol side chain in cysteine enables the formation of Disulfide, disulfide bonds, and often participates in enzymatic reactions as ...
, hydroxylysine, serine
Serine
(symbol Ser or S) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α- amino group (which is in the protonated − form under biological conditions), a carboxyl group (which is in the deprotonated − ...
, threonine
Threonine (symbol Thr or T) is an amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form when dissolved in water), a carboxyl group (which is in the deprotonated −COO− ...
, tyrosine
-Tyrosine or tyrosine (symbol Tyr or Y) or 4-hydroxyphenylalanine is one of the 20 standard amino acids that are used by cells to synthesize proteins. It is a conditionally essential amino acid with a polar side group. The word "tyrosine" is ...
, or tryptophan
Tryptophan (symbol Trp or W)
is an α-amino acid that is used in the biosynthesis of proteins. Tryptophan contains an α-amino group, an α-carboxylic acid group, and a side chain indole, making it a polar molecule with a non-polar aromat ...
resulting in a glycoprotein
Glycoproteins are proteins which contain oligosaccharide (sugar) chains covalently attached to amino acid side-chains. The carbohydrate is attached to the protein in a cotranslational or posttranslational modification. This process is known a ...
. Distinct from glycation
Glycation (non-enzymatic glycosylation) is the covalent bond, covalent attachment of a sugar to a protein, lipid or nucleic acid molecule. Typical sugars that participate in glycation are glucose, fructose, and their derivatives. Glycation is th ...
, which is regarded as a nonenzymatic attachment of sugars.
** ''O''-GlcNAc, addition of ''N''-acetylglucosamine to serine or threonine residues in a β-glycosidic linkage
** polysialylation, addition of polysialic acid (PSA) to neural cell adhesion molecule (NCAM)
* malonylation
* hydroxylation
In chemistry, hydroxylation refers to the installation of a hydroxyl group () into an organic compound. Hydroxylations generate alcohols and phenols, which are very common functional groups. Hydroxylation confers some degree of water-solubility ...
: addition of an oxygen atom to the side-chain of a Pro or Lys residue
* iodination: addition of an iodine atom to the aromatic ring of a tyrosine residue (e.g. in thyroglobulin
Thyroglobulin (Tg) is a 660 kDa, dimeric glycoprotein produced by the follicular cells of the thyroid and used entirely within the thyroid gland. Tg is secreted and accumulated at hundreds of grams per litre in the extracellular compartment ...
)
* nucleotide addition such as ADP-ribosylation
ADP-ribosylation is the addition of one or more ADP-ribose moieties to a protein. It is a reversible post-translational modification that is involved in many cellular processes, including cell signaling, DNA repair, gene regulation and apoptosis ...
* phosphate ester (''O''-linked) or phosphoramidate (''N''-linked) formation
** phosphorylation
In biochemistry, phosphorylation is described as the "transfer of a phosphate group" from a donor to an acceptor. A common phosphorylating agent (phosphate donor) is ATP and a common family of acceptor are alcohols:
:
This equation can be writ ...
, the addition of a phosphate
Phosphates are the naturally occurring form of the element phosphorus.
In chemistry, a phosphate is an anion, salt, functional group or ester derived from a phosphoric acid. It most commonly means orthophosphate, a derivative of orthop ...
group, usually to serine
Serine
(symbol Ser or S) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α- amino group (which is in the protonated − form under biological conditions), a carboxyl group (which is in the deprotonated − ...
, threonine
Threonine (symbol Thr or T) is an amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form when dissolved in water), a carboxyl group (which is in the deprotonated −COO− ...
, and tyrosine
-Tyrosine or tyrosine (symbol Tyr or Y) or 4-hydroxyphenylalanine is one of the 20 standard amino acids that are used by cells to synthesize proteins. It is a conditionally essential amino acid with a polar side group. The word "tyrosine" is ...
(''O''-linked), or histidine
Histidine (symbol His or H) is an essential amino acid that is used in the biosynthesis of proteins. It contains an Amine, α-amino group (which is in the protonated –NH3+ form under Physiological condition, biological conditions), a carboxylic ...
(''N''-linked)
** adenylylation, the addition of an adenylyl moiety, usually to tyrosine
-Tyrosine or tyrosine (symbol Tyr or Y) or 4-hydroxyphenylalanine is one of the 20 standard amino acids that are used by cells to synthesize proteins. It is a conditionally essential amino acid with a polar side group. The word "tyrosine" is ...
(''O''-linked), or histidine
Histidine (symbol His or H) is an essential amino acid that is used in the biosynthesis of proteins. It contains an Amine, α-amino group (which is in the protonated –NH3+ form under Physiological condition, biological conditions), a carboxylic ...
and lysine
Lysine (symbol Lys or K) is an α-amino acid that is a precursor to many proteins. Lysine contains an α-amino group (which is in the protonated form when the lysine is dissolved in water at physiological pH), an α-carboxylic acid group ( ...
(''N''-linked)
** uridylylation, the addition of an uridylyl-group (i.e. uridine monophosphate
Uridine monophosphate (UMP), also known as 5′-uridylic acid ( conjugate base uridylate), is a nucleotide that is used as a monomer in RNA. It is an ester of phosphoric acid with the nucleoside uridine. UMP consists of the phosphate group, th ...
(UMP)), usually to tyrosine
* propionylation
* pyroglutamate formation
* ''S''-glutathionylation
* ''S''-nitrosylation
* ''S''-sulfenylation (''aka'' ''S''-sulphenylation), reversible covalent addition of one oxygen atom to the thiol
In organic chemistry, a thiol (; ), or thiol derivative, is any organosulfur compound of the form , where R represents an alkyl or other organic substituent. The functional group itself is referred to as either a thiol group or a sulfhydryl grou ...
group of a cysteine
Cysteine (; symbol Cys or C) is a semiessential proteinogenic amino acid with the chemical formula, formula . The thiol side chain in cysteine enables the formation of Disulfide, disulfide bonds, and often participates in enzymatic reactions as ...
residue
* ''S''-sulfinylation, normally irreversible covalent addition of two oxygen atoms to the thiol
In organic chemistry, a thiol (; ), or thiol derivative, is any organosulfur compound of the form , where R represents an alkyl or other organic substituent. The functional group itself is referred to as either a thiol group or a sulfhydryl grou ...
group of a cysteine
Cysteine (; symbol Cys or C) is a semiessential proteinogenic amino acid with the chemical formula, formula . The thiol side chain in cysteine enables the formation of Disulfide, disulfide bonds, and often participates in enzymatic reactions as ...
residue
* ''S''-sulfonylation, normally irreversible covalent addition of three oxygen atoms to the thiol
In organic chemistry, a thiol (; ), or thiol derivative, is any organosulfur compound of the form , where R represents an alkyl or other organic substituent. The functional group itself is referred to as either a thiol group or a sulfhydryl grou ...
group of a cysteine
Cysteine (; symbol Cys or C) is a semiessential proteinogenic amino acid with the chemical formula, formula . The thiol side chain in cysteine enables the formation of Disulfide, disulfide bonds, and often participates in enzymatic reactions as ...
residue, resulting in the formation of a cysteic acid residue
* succinylation addition of a succinyl
Succinic acid () is a dicarboxylic acid with the chemical formula (CH2)2(CO2H)2. In living organisms, succinic acid takes the form of an anion, succinate, which has multiple biological roles as a metabolic intermediate being converted into fum ...
group to lysine
Lysine (symbol Lys or K) is an α-amino acid that is a precursor to many proteins. Lysine contains an α-amino group (which is in the protonated form when the lysine is dissolved in water at physiological pH), an α-carboxylic acid group ( ...
* sulfation
Sulfation (sometimes spelled sulphation in British English) is the chemical reaction that entails the addition of SO3 group. In principle, many sulfations would involve reactions of sulfur trioxide (SO3). In practice, most sulfations are effected ...
, the addition of a sulfate group to a tyrosine
-Tyrosine or tyrosine (symbol Tyr or Y) or 4-hydroxyphenylalanine is one of the 20 standard amino acids that are used by cells to synthesize proteins. It is a conditionally essential amino acid with a polar side group. The word "tyrosine" is ...
.
Non-enzymatic modifications ''in vivo''
Examples of non-enzymatic PTMs are glycation, glycoxidation, nitrosylation, oxidation, succination, and lipoxidation. *glycation
Glycation (non-enzymatic glycosylation) is the covalent bond, covalent attachment of a sugar to a protein, lipid or nucleic acid molecule. Typical sugars that participate in glycation are glucose, fructose, and their derivatives. Glycation is th ...
, the addition of a sugar molecule to a protein without the controlling action of an enzyme.
* carbamylation the addition of Isocyanic acid
Isocyanic acid is a chemical compound with the structural formula HNCO, which is often written as . It is a colourless, volatile and poisonous gas, condensing at 23.5 °C. It is the predominant tautomer and an isomer of cyanic acid ''( ...
to a protein's N-terminus or the side-chain of Lys.
* carbonylation
In chemistry, carbonylation refers to reactions that introduce carbon monoxide (CO) into organic and inorganic substrates. Carbon monoxide is abundantly available and conveniently reactive, so it is widely used as a reactant in industrial chemis ...
the addition of carbon monoxide to other organic/inorganic compounds.
* spontaneous isopeptide bond formation, as found in many surface proteins of Gram-positive bacteria
In bacteriology, gram-positive bacteria are bacteria that give a positive result in the Gram stain test, which is traditionally used to quickly classify bacteria into two broad categories according to their type of cell wall.
The Gram stain ...
.
Non-enzymatic additions ''in vitro''
* biotinylation: covalent attachment of a biotin moiety using a biotinylation reagent, typically for the purpose of labeling a protein. * carbamylation: the addition of isocyanic acid to a protein's N-terminus or the side-chain of Lys or Cys residues, typically resulting from exposure to urea solutions. * oxidation: addition of one or more oxygen atoms to a susceptible side-chain, principally of Met, Trp, His or Cys residues. Formation ofdisulfide
In chemistry, a disulfide (or disulphide in British English) is a compound containing a functional group or the anion. The linkage is also called an SS-bond or sometimes a disulfide bridge and usually derived from two thiol groups.
In inorg ...
bonds between Cys residues.
* pegylation
PEGylation (or pegylation) is the process of both covalent and non-covalent attachment or amalgamation of polyethylene glycol (PEG, in pharmacy called macrogol) polymer chains to molecules and macrostructures, such as a drug, therapeutic protein ...
: covalent attachment of polyethylene glycol
Polyethylene glycol (PEG; ) is a polyether compound derived from petroleum with many applications, from industrial manufacturing to medicine. PEG is also known as polyethylene oxide (PEO) or polyoxyethylene (POE), depending on its molecular wei ...
(PEG) using a pegylation reagent, typically to the N-terminus or the side-chains of Lys residues. Pegylation is used to improve the efficacy of protein pharmaceuticals.
Conjugation with other proteins or peptides
*ubiquitin
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 19 ...
ation, the covalent
A covalent bond is a chemical bond that involves the sharing of electrons to form electron pairs between atoms. These electron pairs are known as shared pairs or bonding pairs. The stable balance of attractive and repulsive forces between atom ...
linkage to the protein ubiquitin.
* SUMOylation
In molecular biology, SUMO (Small Ubiquitin-like Modifier) proteins are a family of small proteins that are covalently attached to and detached from other proteins in cells to modify their function. This process is called SUMOylation (pronounced ...
, the covalent linkage to the SUMO protein
In molecular biology, SUMO (Small Ubiquitin-like Modifier) proteins are a Protein family, family of small proteins that are covalent bond, covalently attached to and detached from other proteins in cell (biology), cells to modify their function. T ...
(small ubiquitin-related modifier)
* neddylation, the covalent linkage to the Nedd protein
* ISGylation, the covalent linkage to the ISG15 protein (interferon-stimulated gene 15)
* pupylation, the covalent linkage to the prokaryotic ubiquitin-like protein
Chemical modification of amino acids
* citrullination, or deimination, the conversion ofarginine
Arginine is the amino acid with the formula (H2N)(HN)CN(H)(CH2)3CH(NH2)CO2H. The molecule features a guanidinium, guanidino group appended to a standard amino acid framework. At physiological pH, the carboxylic acid is deprotonated (−CO2−) a ...
to citrulline
The organic compound citrulline is an α-amino acid. Its name is derived from '' citrullus'', the Latin word for watermelon. Although named and described by gastroenterologists since the late 19th century, it was first isolated from watermelon in ...
* deamidation
Deamidation is a chemical reaction in which an amide functional group in the side chain of the amino acids asparagine or glutamine is removed or converted to another functional group. Typically, asparagine is converted to aspartic acid or isoasp ...
, the conversion of glutamine
Glutamine (symbol Gln or Q) is an α-amino acid that is used in the biosynthesis of proteins. Its side chain is similar to that of glutamic acid, except the carboxylic acid group is replaced by an amide. It is classified as a charge-neutral ...
to glutamic acid
Glutamic acid (symbol Glu or E; known as glutamate in its anionic form) is an α- amino acid that is used by almost all living beings in the biosynthesis of proteins. It is a non-essential nutrient for humans, meaning that the human body can ...
or asparagine
Asparagine (symbol Asn or N) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form under biological conditions), an α-carboxylic acid group (which is in the depro ...
to aspartic acid
Aspartic acid (symbol Asp or D; the ionic form is known as aspartate), is an α-amino acid that is used in the biosynthesis of proteins. The L-isomer of aspartic acid is one of the 22 proteinogenic amino acids, i.e., the building blocks of protei ...
* eliminylation, the conversion to an alkene
In organic chemistry, an alkene, or olefin, is a hydrocarbon containing a carbon–carbon double bond. The double bond may be internal or at the terminal position. Terminal alkenes are also known as Alpha-olefin, α-olefins.
The Internationa ...
by beta-elimination of phosphothreonine and phosphoserine, or dehydration
In physiology, dehydration is a lack of total body water that disrupts metabolic processes. It occurs when free water loss exceeds intake, often resulting from excessive sweating, health conditions, or inadequate consumption of water. Mild deh ...
of threonine
Threonine (symbol Thr or T) is an amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form when dissolved in water), a carboxyl group (which is in the deprotonated −COO− ...
and serine
Serine
(symbol Ser or S) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α- amino group (which is in the protonated − form under biological conditions), a carboxyl group (which is in the deprotonated − ...
Structural changes
* disulfide bridges, the covalent linkage of twocysteine
Cysteine (; symbol Cys or C) is a semiessential proteinogenic amino acid with the chemical formula, formula . The thiol side chain in cysteine enables the formation of Disulfide, disulfide bonds, and often participates in enzymatic reactions as ...
amino acids
* lysine-cysteine bridges, the covalent linkage of 1 lysine and 1 or 2 cystine residues via an oxygen atom (NOS and SONOS bridges)
* proteolytic cleavage, cleavage of a protein at a peptide bond
* isoaspartate formation, via the cyclisation of asparagine or aspartic acid amino-acid residues
* racemization
** of serine
Serine
(symbol Ser or S) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α- amino group (which is in the protonated − form under biological conditions), a carboxyl group (which is in the deprotonated − ...
by protein-serine epimerase
** of alanine in dermorphin, a frog opioid peptide
** of methionine
Methionine (symbol Met or M) () is an essential amino acid in humans.
As the precursor of other non-essential amino acids such as cysteine and taurine, versatile compounds such as SAM-e, and the important antioxidant glutathione, methionine play ...
in deltorphin, also a frog opioid peptide
* protein splicing, self-catalytic removal of inteins analogous to mRNA processing
Statistics
Common PTMs by frequency
In 2011, statistics of each post-translational modification experimentally and putatively detected have been compiled using proteome-wide information from the Swiss-Prot database. The 10 most common experimentally found modifications were as follows:Common PTMs by residue
Some common post-translational modifications to specific amino-acid residues are shown below. Modifications occur on the side-chain unless indicated otherwise.Databases and tools
 Protein sequences contain sequence motifs that are recognized by modifying enzymes, and which can be documented or predicted in PTM databases. With the large number of different modifications being discovered, there is a need to document this sort of information in databases. PTM information can be collected through experimental means or predicted from high-quality, manually curated data. Numerous databases have been created, often with a focus on certain taxonomic groups (e.g. human proteins) or other features.
Protein sequences contain sequence motifs that are recognized by modifying enzymes, and which can be documented or predicted in PTM databases. With the large number of different modifications being discovered, there is a need to document this sort of information in databases. PTM information can be collected through experimental means or predicted from high-quality, manually curated data. Numerous databases have been created, often with a focus on certain taxonomic groups (e.g. human proteins) or other features.
List of resources
PhosphoSitePlus
– A database of comprehensive information and tools for the study of mammalian protein post-translational modification * ProteomeScout – A database of proteins and post-translational modifications experimentally * Human Protein Reference Database – A database for different modifications and understand different proteins, their class, and function/process related to disease causing proteins * PROSITE – A database of Consensus patterns for many types of PTM's including sites * Protein Information Resource, RESID – A database consisting of a collection of annotations and structures for PTMs.
iPTMnet
– A database that integrates PTM information from several knowledgbases and text mining results. * dbPTM – A database that shows different PTM's and information regarding their chemical components/structures and a frequency for amino acid modified site
Uniprot
has PTM information although that may be less comprehensive than in more specialized databases.

The ''O''-GlcNAc Database
- A curated database for protein O-GlcNAcylation and referencing more than 14 000 protein entries and 10 000 ''O''-GlcNAc sites.
Tools
List of software for visualization of proteins and their PTMs * PyMOL – introduce a set of common PTM's into protein models * AWESOME – Interactive tool to see the role of single nucleotide polymorphisms to PTM's * UCSF Chimera, Chimera – Interactive Database to visualize moleculesCase examples
* Cleavage and formation of disulfide bridges during the production ofinsulin
Insulin (, from Latin ''insula'', 'island') is a peptide hormone produced by beta cells of the pancreatic islets encoded in humans by the insulin (''INS)'' gene. It is the main Anabolism, anabolic hormone of the body. It regulates the metabol ...
* PTM of histones as regulation of Transcription (genetics), transcription: RNA polymerase control by chromatin structure
* PTM of RNA polymerase II as regulation of transcription
* Cleavage of polypeptide chains as crucial for lectin specificity
* Influence of Ni(II) in the Acetylation of Histones H4 Protein
See also
* Protein targeting * Post-translational regulationReferences
External links
Controlled vocabulary of post-translational modifications
in Uniprot
List of posttranslational modifications in ExPASy
Browse SCOP domains by PTM
— from the dcGO database
Overview and description of commonly used post-translational modification detection techniques
{{DEFAULTSORT:Posttranslational Modification Gene expression Protein structure Protein biosynthesis Post-translational modification, Cell biology