|
Pupylation
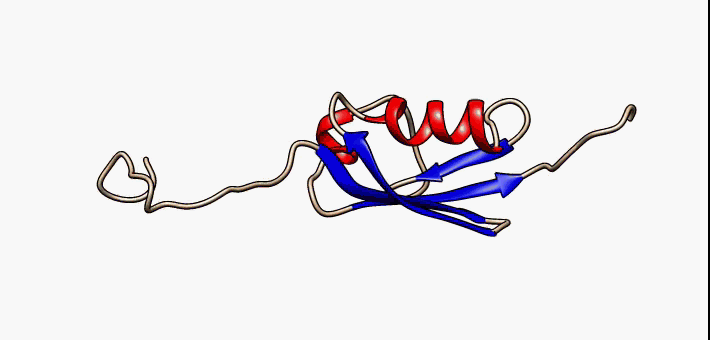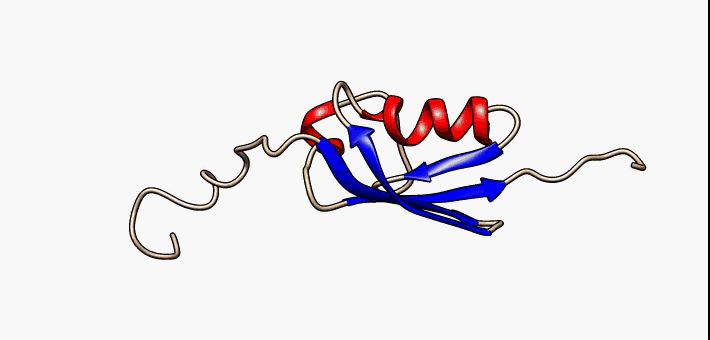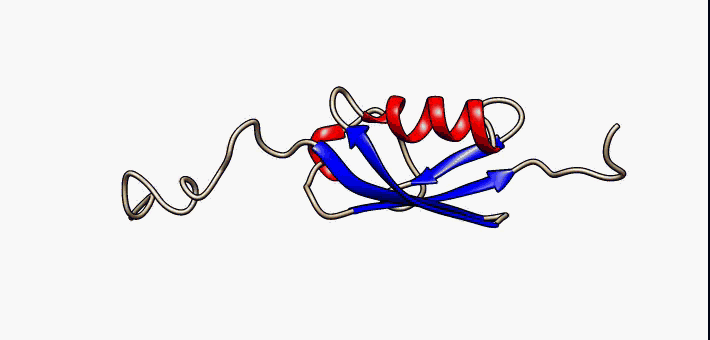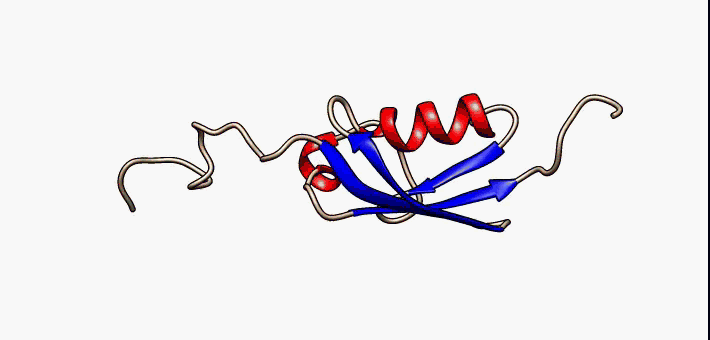
Prokaryotic ubiquitin-like protein (Pup) is a functional analog of ubiquitin found in the prokaryote ''Mycobacterium tuberculosis''. Like ubiquitin, Pup serves to direct proteins to the proteasome for degradation in the Pup-proteasome system (PPS). However, the enzymology of ubiquitylation and pupylation is different, owing to their distinct evolutionary origins. In contrast to the three-step reaction of ubiquitylation, pupylation requires only two steps, and thus only two enzymes are involved in pupylation. The enzymes involved in pupylation are descended from glutamine synthetase. Similar to ubiquitin, Pup is attached to specific lysine residues of substrate proteins by isopeptide bonds; this is called pupylation. It is then recognized by the protein Mycobacterium proteasomal ATPase (Mpa), in a mechanism that induces folding of Pup. Mpa delivers the substrate protein to the proteasome for degradation by coupling of ATP hydrolysis. The discovery of Pup indicates that like euka ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
UBact Family
Prokaryotic ubiquitin-like protein (Pup) is a functional analog of ubiquitin found in the prokaryote ''Mycobacterium tuberculosis''. Like ubiquitin, Pup serves to direct proteins to the proteasome for degradation in the Pup-proteasome system (PPS). However, the enzymology of ubiquitylation and pupylation is different, owing to their distinct evolutionary origins. In contrast to the three-step reaction of ubiquitylation, pupylation requires only two steps, and thus only two enzymes are involved in pupylation. The enzymes involved in pupylation are descended from glutamine synthetase. Similar to ubiquitin, Pup is attached to specific lysine residues of substrate proteins by isopeptide bonds; this is called pupylation. It is then recognized by the protein Mycobacterium proteasomal ATPase (Mpa), in a mechanism that induces folding of Pup. Mpa delivers the substrate protein to the proteasome for degradation by coupling of ATP hydrolysis. The discovery of Pup indicates that like eukary ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pupylation
Prokaryotic ubiquitin-like protein (Pup) is a functional analog of ubiquitin found in the prokaryote ''Mycobacterium tuberculosis''. Like ubiquitin, Pup serves to direct proteins to the proteasome for degradation in the Pup-proteasome system (PPS). However, the enzymology of ubiquitylation and pupylation is different, owing to their distinct evolutionary origins. In contrast to the three-step reaction of ubiquitylation, pupylation requires only two steps, and thus only two enzymes are involved in pupylation. The enzymes involved in pupylation are descended from glutamine synthetase. Similar to ubiquitin, Pup is attached to specific lysine residues of substrate proteins by isopeptide bonds; this is called pupylation. It is then recognized by the protein Mycobacterium proteasomal ATPase (Mpa), in a mechanism that induces folding of Pup. Mpa delivers the substrate protein to the proteasome for degradation by coupling of ATP hydrolysis. The discovery of Pup indicates that like euka ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin Bacterial
Prokaryotic ubiquitin-like protein (Pup) is a functional analog of ubiquitin found in the prokaryote ''Mycobacterium tuberculosis''. Like ubiquitin, Pup serves to direct proteins to the proteasome for degradation in the Pup-proteasome system (PPS). However, the enzymology of ubiquitylation and pupylation is different, owing to their distinct evolutionary origins. In contrast to the three-step reaction of ubiquitylation, pupylation requires only two steps, and thus only two enzymes are involved in pupylation. The enzymes involved in pupylation are descended from glutamine synthetase. Similar to ubiquitin, Pup is attached to specific lysine residues of substrate proteins by isopeptide bonds; this is called pupylation. It is then recognized by the protein Mycobacterium proteasomal ATPase (Mpa), in a mechanism that induces folding of Pup. Mpa delivers the substrate protein to the proteasome for degradation by coupling of ATP hydrolysis. The discovery of Pup indicates that like eukary ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A. The addition of ubiquitin to a substrate protein is called ubiquitylation (or, alternatively, ubiquitination or ubiquitinylation). Ubiquitylation affects proteins in many ways: it can mark them for degradation via the proteasome, alter their cellular location, affect their activity, and promote or prevent protein interactions. Ubiquitylation involves three main steps: activation, conjugation, and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), respectively. The result of this sequential cascade is to bind ubiquitin to lysine residues on the protein substrate via an isopeptide bond, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A. The addition of ubiquitin to a substrate protein is called ubiquitylation (or, alternatively, ubiquitination or ubiquitinylation). Ubiquitylation affects proteins in many ways: it can mark them for degradation via the proteasome, alter their cellular location, affect their activity, and promote or prevent protein interactions. Ubiquitylation involves three main steps: activation, conjugation, and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), respectively. The result of this sequential cascade is to bind ubiquitin to lysine residues on the protein substrate via an isopeptide bond, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitylation
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A. The addition of ubiquitin to a substrate protein is called ubiquitylation (or, alternatively, ubiquitination or ubiquitinylation). Ubiquitylation affects proteins in many ways: it can mark them for degradation via the proteasome, alter their cellular location, affect their activity, and promote or prevent protein interactions. Ubiquitylation involves three main steps: activation, conjugation, and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), respectively. The result of this sequential cascade is to bind ubiquitin to lysine residues on the protein substrate via an isopeptide bond, cyst ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Actinomycetota
The ''Actinomycetota'' (or ''Actinobacteria'') are a phylum of all gram-positive bacteria. They can be terrestrial or aquatic. They are of great economic importance to humans because agriculture and forests depend on their contributions to soil systems. In soil they help to decompose the organic matter of dead organisms so the molecules can be taken up anew by plants. While this role is also played by fungi, ''Actinomycetota'' are much smaller and likely do not occupy the same ecological niche. In this role the colonies often grow extensive mycelia, like a fungus would, and the name of an important order of the phylum, '' Actinomycetales'' (the actinomycetes), reflects that they were long believed to be fungi. Some soil actinomycetota (such as ''Frankia'') live symbiotically with the plants whose roots pervade the soil, fixing nitrogen for the plants in exchange for access to some of the plant's saccharides. Other species, such as many members of the genus '' Mycobacterium'', ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aaron Ciechanover
Aaron Ciechanover ( ; he, אהרן צ'חנובר; born October 1, 1947) is an Israeli biologist who won the Nobel Prize in Chemistry for characterizing the method that cells use to degrade and recycle proteins using ubiquitin. Biography Early life Ciechanover was born in Haifa, British Mandate of Palestine on 1 October 1947. He is the son of Bluma (Lubashevsky), a teacher of English, and Yitzhak Ciechanover, an office worker. His mother and father supported the Zionist movement and immigrated to Israel from Poland in the 1920s. Education He earned a master's degree in science in 1971 and graduated from Hadassah Medical School in Jerusalem in 1974. He received his doctorate in biochemistry in 1981 from the Technion – Israel Institute of Technology in Haifa before conducting postdoctoral research in the laboratory of Harvey Lodish at the Whitehead Institute at MIT from 1981 to 1984. Recent Ciechanover is currently a Technion Distinguished Research Professor in the Rut ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Israel
Israel (; he, יִשְׂרָאֵל, ; ar, إِسْرَائِيل, ), officially the State of Israel ( he, מְדִינַת יִשְׂרָאֵל, label=none, translit=Medīnat Yīsrāʾēl; ), is a country in Western Asia. It is situated on the southeastern shore of the Mediterranean Sea and the northern shore of the Red Sea, and shares borders with Lebanon to the north, Syria to the northeast, Jordan to the east, and Egypt to the southwest. Israel also is bordered by the Palestinian territories of the West Bank and the Gaza Strip to the east and west, respectively. Tel Aviv is the economic and technological center of the country, while its seat of government is in its proclaimed capital of Jerusalem, although Israeli sovereignty over East Jerusalem is unrecognized internationally. The land held by present-day Israel witnessed some of the earliest human occupations outside Africa and was among the earliest known sites of agriculture. It was inhabited by the Canaanites ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intrinsically Disordered Proteins
In molecular biology, an intrinsically disordered protein (IDP) is a protein that lacks a fixed or ordered three-dimensional structure, typically in the absence of its macromolecular interaction partners, such as other proteins or RNA. IDPs range from fully unstructured to partially structured and include random coil, molten globule-like aggregates, or flexible linkers in large multi-domain proteins. They are sometimes considered as a separate class of proteins along with globular, fibrous and membrane proteins. IDPs are a very large and functionally important class of proteins and their discovery has disproved the idea that three-dimensional structures of proteins must be fixed to accomplish their biological functions. For example, IDPs have been identified to participate in weak multivalent interactions that are highly cooperative and dynamic, lending them importance in DNA regulation and in cell signaling. Many IDPs can also adopt a fixed three-dimensional structure after bi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryotes
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacteria and Archaea (both prokaryotes) make up the other two domains. The eukaryotes are usually now regarded as having emerged in the Archaea or as a sister of the Asgard archaea. This implies that there are only two domains of life, Bacteria and Archaea, with eukaryotes incorporated among archaea. Eukaryotes represent a small minority of the number of organisms, but, due to their generally much larger size, their collective global biomass is estimated to be about equal to that of prokaryotes. Eukaryotes emerged approximately 2.3–1.8 billion years ago, during the Proterozoic eon, likely as flagellated phagotrophs. Their name comes from the Greek εὖ (''eu'', "well" or "good") and κάρυον (''karyon'', "nut" or "kernel"). Eu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |