|
PreQ1-III Riboswitch
PreQ1-III riboswitches are a class of riboswitches that bind pre-queuosine1 (PreQ1), a precursor to the modified nucleoside queuosine. PreQ1-III riboswitches are the third class of riboswitches to be discovered that sense this ligand, and are structurally distinct from preQ1-I and preQ1-II riboswitches. Most sequenced examples of preQ1-III riboswitches are obtained from uncultivated metagenome samples, but the few examples in cultivated organisms are present in strains that are known to or suspected to be ''Faecalibacterium prausnitzii'', a species of Gram-positive Clostridia The Clostridia are a highly polyphyletic class of Bacillota, including '' Clostridium'' and other similar genera. They are distinguished from the Bacilli by lacking aerobic respiration. They are obligate anaerobes and oxygen is toxic to them. Sp .... Known examples of preQ1-III riboswitches are found upstream of ''queT'' genes, which are expected to encode transporters of a queuosine derivative. The othe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Riboswitch
In molecular biology, a riboswitch is a regulatory segment of a messenger RNA molecule that binds a small molecule, resulting in a change in production of the proteins encoded by the mRNA. Thus, an mRNA that contains a riboswitch is directly involved in regulating its own activity, in response to the concentrations of its effector molecule. The discovery that modern organisms use RNA to bind small molecules, and discriminate against closely related analogs, expanded the known natural capabilities of RNA beyond its ability to code for proteins, catalyze reactions, or to bind other RNA or protein macromolecules. The original definition of the term "riboswitch" specified that they directly sense small-molecule metabolite concentrations. Although this definition remains in common use, some biologists have used a broader definition that includes other cis-regulatory RNAs. However, this article will discuss only metabolite-binding riboswitches. Most known riboswitches occur in bac ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Queuosine
Queuosine is a modified nucleoside that is present in certain tRNAs in bacteria and eukaryotes. It contains the nucleobase queuine. Originally identified in ''E. coli'', queuosine was found to occupy the first anticodon position of tRNAs for histidine, aspartic acid, asparagine and tyrosine. The first anticodon position pairs with the third " wobble" position in codons, and queuosine improves accuracy of translation compared to guanosine. Synthesis of queuosine begins with GTP. In bacteria, three structurally unrelated classes of riboswitch are known to regulate genes that are involved in the synthesis or transport of pre-queuosine1, a precursor to queuosine: PreQ1-I riboswitches, PreQ1-II riboswitches and PreQ1-III riboswitch PreQ1-III riboswitches are a class of riboswitches that bind pre-queuosine1 (PreQ1), a precursor to the modified nucleoside queuosine. PreQ1-III riboswitches are the third class of riboswitches to be discovered that sense this ligand, and are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PreQ1 Riboswitch
The PreQ1-I riboswitch is a cis-acting element identified in bacteria which regulates expression of genes involved in biosynthesis of the nucleoside queuosine (Q) from GTP. PreQ1 (pre-queuosine1) is an intermediate in the queuosine pathway, and preQ1 riboswitch, as a type of riboswitch, is an RNA element that binds preQ1. The preQ1 riboswitch is distinguished by its unusually small aptamer, compared to other riboswitches. Its atomic-resolution three-dimensional structure has been determined, with thPDB ID 2L1V PreQ1 classification Three subcategories of the PreQ1 riboswitch exist: preQ1-I, preQ1-II, and preQ1-III. PreQ1-I has a distinctly small aptamer, ranging from 25 to 45 nucleotides long, compared to the structures of PreQ1-II riboswitch and preQ1-III riboswitch. PreQ1-II riboswitch, only found in '' Lactobacillales'', has a larger and more complex consensus sequence and structure than preQ1-I riboswitch, with an average of 58 nucleotides composing its aptamer, which forms ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PreQ1-II Riboswitch
PreQ1-II riboswitches form a class of riboswitches that specifically bind pre-queuosine1 (PreQ1), a precursor of the modified nucleoside queuosine. They are found in certain species of ''Streptococcus'' and ''Lactococcus'', and were originally identified as a conserved RNA secondary structure called the "COG4708 motif". All known members of this riboswitch class appear to control members of COG4708 genes. These genes are predicted to encode membrane-bound proteins and have been proposed to be a transporter of preQ1, or a related metabolite, based on their association with preQ1-binding riboswitches. PreQ1-II riboswitches have no apparent similarities in sequence or structure to preQ1-I riboswitches, a previously discovered class of preQ1-binding riboswitches. PreQ1 thus joins S-adenosylmethionine ''S''-Adenosyl methionine (SAM), also known under the commercial names of SAMe, SAM-e, or AdoMet, is a common cosubstrate involved in methyl group transfers, transsulfuration, a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
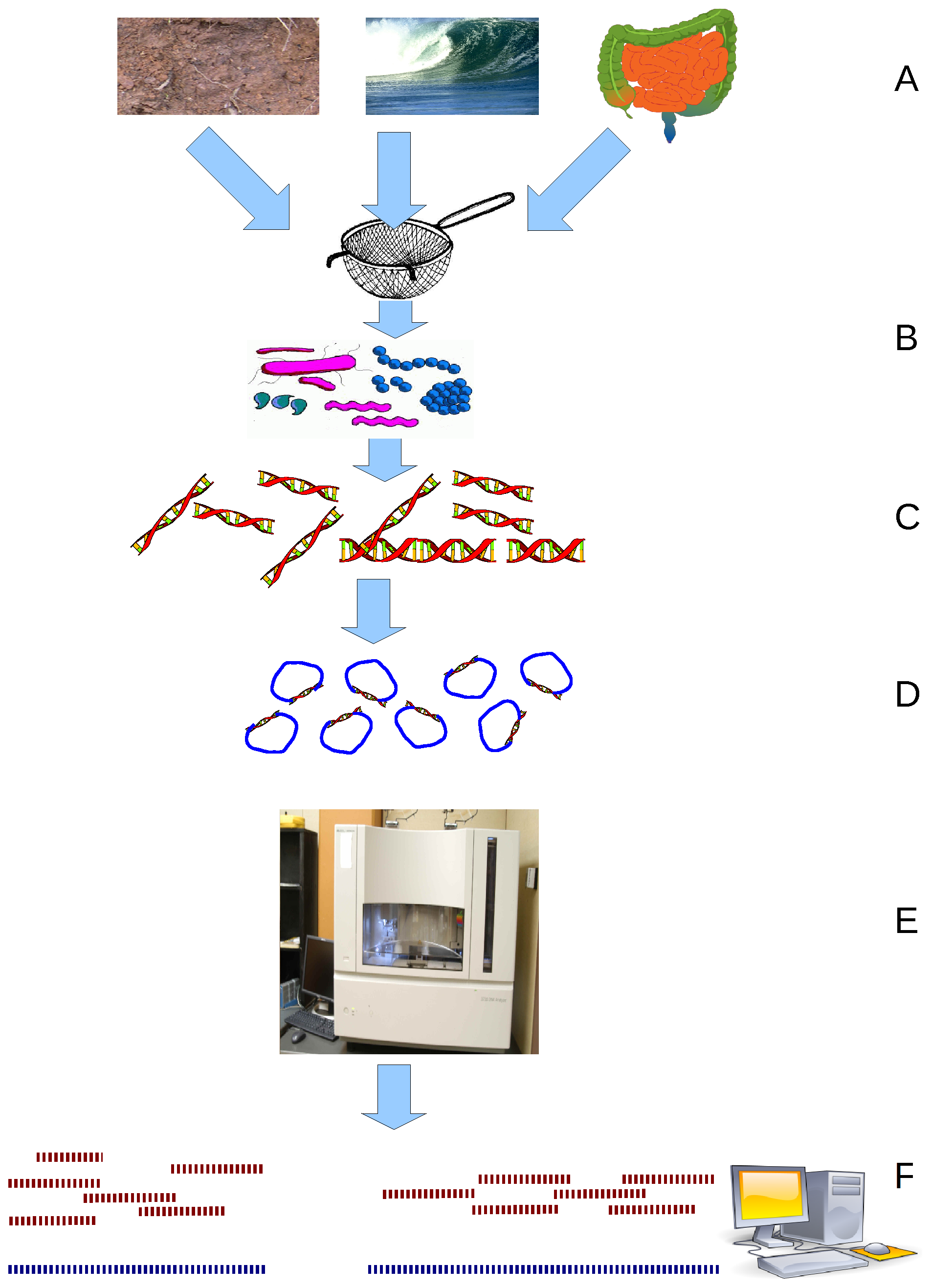
Metagenome
Metagenomics is the study of genetic material recovered directly from environmental or clinical samples by a method called sequencing. The broad field may also be referred to as environmental genomics, ecogenomics, community genomics or microbiomics. While traditional microbiology and microbial genome sequencing and genomics rely upon cultivated clonal cultures, early environmental gene sequencing cloned specific genes (often the 16S rRNA gene) to produce a profile of diversity in a natural sample. Such work revealed that the vast majority of microbial biodiversity had been missed by cultivation-based methods. Because of its ability to reveal the previously hidden diversity of microscopic life, metagenomics offers a powerful lens for viewing the microbial world that has the potential to revolutionize understanding of the entire living world. As the price of DNA sequencing continues to fall, metagenomics now allows microbial ecology to be investigated at a much greater scale ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Faecalibacterium Prausnitzii
''Faecalibacterium'' is a genus of bacteria. Its sole known species, ''Faecalibacterium prausnitzii'' is gram-positive, mesophilic, rod-shaped, anaerobic and is one of the most abundant and important commensal bacteria of the human gut microbiota. It is non-spore forming and non-motile. These bacteria produce butyrate and other short-chain fatty acids through the fermentation of dietary fiber. History Formerly considered to be a member of ''Fusobacterium'', the bacterium is named in honor of German bacteriologist Otto Prausnitz. In 2002, it was proposed to be reclassified as its own genus, ''Faecalibacterium'', containing the species ''Faecalibacterium prausnitzii'', as phylogenetic analysis from isolates showed it to be only distantly related to ''Fusobacterium'', and a closer member of Clostridium cluster IV. Genetics ''Faecalibacterium prausnitzii'' has a genome 2,868,932 bp long and has a GC-content of 56.9%. The bacterium has been found to have 2,707 coding sequences, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gram-positive
In bacteriology, gram-positive bacteria are bacteria that give a positive result in the Gram stain test, which is traditionally used to quickly classify bacteria into two broad categories according to their type of cell wall. Gram-positive bacteria take up the crystal violet stain used in the test, and then appear to be purple-coloured when seen through an optical microscope. This is because the thick peptidoglycan layer in the bacterial cell wall retains the stain after it is washed away from the rest of the sample, in the decolorization stage of the test. Conversely, gram-negative bacteria cannot retain the violet stain after the decolorization step; alcohol used in this stage degrades the outer membrane of gram-negative cells, making the cell wall more porous and incapable of retaining the crystal violet stain. Their peptidoglycan layer is much thinner and sandwiched between an inner cell membrane and a bacterial outer membrane, causing them to take up the counterstain (saf ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Clostridia
The Clostridia are a highly polyphyletic class of Bacillota, including '' Clostridium'' and other similar genera. They are distinguished from the Bacilli by lacking aerobic respiration. They are obligate anaerobes and oxygen is toxic to them. Species of the class ''Clostridia'' are often but not always Gram-positive (see ''Halanaerobium'') and have the ability to form spores. Studies show they are not a monophyletic group, and their relationships are not entirely certain. Currently, most are placed in a single order called Clostridiales, but this is not a natural group and is likely to be redefined in the future. Most species of the genus ''Clostridium'' are saprophytic organisms that ferment plant polysaccharides and are found in many places in the environment, most notably the soil. However, the genus does contain some human pathogens (outlined below). The toxins produced by certain members of the genus ''Clostridium'' are among the most dangerous known. Examples are tetanus ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cis-regulatory RNA Elements
''Cis-acting replication elements'' bring together the 5′ and 3′ ends during replication of positive-sense single-stranded RNA viruses (for example Picornavirus, Flavivirus, coronavirus, togaviruses, Hepatitis C virus) and double-stranded RNA viruses (for example rotavirus and reovirus). See also *Cis-regulatory element *List of cis-regulatory RNA elements *Enterovirus cis-acting replication element and Enterovirus 5′ cloverleaf cis-acting replication element *Cardiovirus cis-acting replication element (CRE) *Coronavirus SL-III cis-acting replication element (CRE) *Rotavirus cis-acting replication element *Hepatitis C virus cis-acting replication element *Flavivirus 3′ UTR cis-acting replication element (CRE) *Potato virus X cis-acting regulatory element *Human rhinovirus internal cis-acting regulatory element (CRE) Human rhinovirus internal cis-acting regulatory element (CRE) is a CRE from the human rhinoviruses. The CRE is located within the genome segment encoding the c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |

.png)