|
OrthoDB Logo
OrthoDB presents a catalog of orthologous protein-coding genes across vertebrates, arthropods, fungi, plants, and bacteria. Orthology refers to the last common ancestor of the species under consideration, and thus OrthoDB explicitly delineates orthologs at each major radiation along the species phylogeny. The database of orthologs presents available protein descriptors, together with Gene Ontology and InterPro attributes, which serve to provide general descriptive annotations of the orthologous groups, and facilitate comprehensive orthology database querying. OrthoDB also provides computed evolutionary traits of orthologs, such as gene duplicability and loss profiles, divergence rates, sibling groups, and gene intron-exon architectures. In comparative genomics, the importance of scale cannot be underestimated. As gene orthology delineation requires specific expertise and considerable computational resources, scale is something that individual non-specialist research groups canno ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
OrthoDB Logo
OrthoDB presents a catalog of orthologous protein-coding genes across vertebrates, arthropods, fungi, plants, and bacteria. Orthology refers to the last common ancestor of the species under consideration, and thus OrthoDB explicitly delineates orthologs at each major radiation along the species phylogeny. The database of orthologs presents available protein descriptors, together with Gene Ontology and InterPro attributes, which serve to provide general descriptive annotations of the orthologous groups, and facilitate comprehensive orthology database querying. OrthoDB also provides computed evolutionary traits of orthologs, such as gene duplicability and loss profiles, divergence rates, sibling groups, and gene intron-exon architectures. In comparative genomics, the importance of scale cannot be underestimated. As gene orthology delineation requires specific expertise and considerable computational resources, scale is something that individual non-specialist research groups canno ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
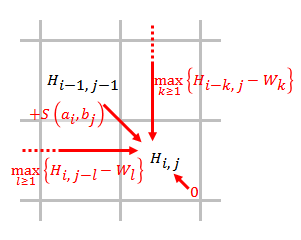
Smith–Waterman Algorithm
The Smith–Waterman algorithm performs local sequence alignment; that is, for determining similar regions between two strings of nucleic acid sequences or protein sequences. Instead of looking at the entire sequence, the Smith–Waterman algorithm compares segments of all possible lengths and optimizes the similarity measure. The algorithm was first proposed by Temple F. Smith and Michael S. Waterman in 1981. Like the Needleman–Wunsch algorithm, of which it is a variation, Smith–Waterman is a dynamic programming algorithm. As such, it has the desirable property that it is guaranteed to find the optimal local alignment with respect to the scoring system being used (which includes the substitution matrix and the gap-scoring scheme). The main difference to the Needleman–Wunsch algorithm is that negative scoring matrix cells are set to zero, which renders the (thus positively scoring) local alignments visible. Traceback procedure starts at the highest scoring matrix cell ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biological Databases
Biological databases are libraries of biological sciences, collected from scientific experiments, published literature, high-throughput experiment technology, and computational analysis. They contain information from research areas including genomics, proteomics, metabolomics, microarray gene expression, and phylogenetics. Information contained in biological databases includes gene function, structure, localization (both cellular and chromosomal), clinical effects of mutations as well as similarities of biological sequences and structures. Biological databases can be classified by the kind of data they collect (see below). Broadly, there are molecular databases (for sequences, molecules, etc.), functional databases (for physiology, enzyme activities, phenotypes, ecology etc), taxonomic databases (for species and other taxonomic ranks), images and other media, or specimens (for museum collections etc.) Databases are important tools in assisting scientists to analyze and explain a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Biological Databases
Biological databases are stores of biological information. The journal ''Nucleic Acids Research'' regularly publishes special issues on biological databases and has a list of such databases. The 2018 issue has a list of about 180 such databases and updates to previously described databasesOmics Discovery Indexcan be used to browse and search several biological databases. Meta databases Meta databases are databases of databases that collect data about data to generate new data. They are capable of merging information from different sources and making it available in a new and more convenient form, or with an emphasis on a particular disease or organism. etadatabase is a database model for metadata management, global query of independent database, and distributed data processing. The word metadatabase is an addition to the dictionary originally ,metadata was only common term referring simply to ''data about data '' such a tags ,keywords, and markup headers. * ConsensusPathDB: a m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylogeny
A phylogenetic tree (also phylogeny or evolutionary tree Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA.) is a branching diagram or a tree showing the evolutionary relationships among various biological species or other entities based upon similarities and differences in their physical or genetic characteristics. All life on Earth is part of a single phylogenetic tree, indicating common ancestry. In a ''rooted'' phylogenetic tree, each node with descendants represents the inferred most recent common ancestor of those descendants, and the edge lengths in some trees may be interpreted as time estimates. Each node is called a taxonomic unit. Internal nodes are generally called hypothetical taxonomic units, as they cannot be directly observed. Trees are useful in fields of biology such as bioinformatics, systematics, and phylogenetics. ''Unrooted'' trees illustrate only the relatedness of the leaf nodes and do not require the ancestral root to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homology (biology)
In biology, homology is similarity due to shared ancestry between a pair of structures or genes in different taxa. A common example of homologous structures is the forelimbs of vertebrates, where the wings of bats and birds, the arms of primates, the front flippers of whales and the forelegs of four-legged vertebrates like dogs and crocodiles are all derived from the same ancestral tetrapod structure. Evolutionary biology explains homologous structures adapted to different purposes as the result of descent with modification from a common ancestor. The term was first applied to biology in a non-evolutionary context by the anatomist Richard Owen in 1843. Homology was later explained by Charles Darwin's theory of evolution in 1859, but had been observed before this, from Aristotle onwards, and it was explicitly analysed by Pierre Belon in 1555. In developmental biology, organs that developed in the embryo in the same manner and from similar origins, such as from matching prim ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FlyBase
FlyBase is an online bioinformatics database and the primary repository of genetic and molecular data for the insect family Drosophilidae. For the most extensively studied species and model organism, ''Drosophila melanogaster'', a wide range of data are presented in different formats. Information in FlyBase originates from a variety of sources ranging from large-scale genome projects to the primary research literature. These data types include mutant phenotypes; molecular characterization of mutant alleles; and other deviations, cytological maps, wild-type expression patterns, anatomical images, transgenic constructs and insertions, sequence-level gene models, and molecular classification of gene product functions. Query tools allow navigation of FlyBase through DNA or protein sequence, by gene or mutant name, or through terms from the several ontologies used to capture functional, phenotypic, and anatomical data. The database offers several different query tools in order to prov ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
National Center For Biotechnology Information
The National Center for Biotechnology Information (NCBI) is part of the United States National Library of Medicine (NLM), a branch of the National Institutes of Health (NIH). It is approved and funded by the government of the United States. The NCBI is located in Bethesda, Maryland, and was founded in 1988 through legislation sponsored by US Congressman Claude Pepper. The NCBI houses a series of databases relevant to biotechnology and biomedicine and is an important resource for bioinformatics tools and services. Major databases include GenBank for DNA sequences and PubMed, a bibliographic database for biomedical literature. Other databases include the NCBI Epigenomics database. All these databases are available online through the Entrez search engine. NCBI was directed by David Lipman, one of the original authors of the BLAST sequence alignment program and a widely respected figure in bioinformatics. GenBank NCBI had responsibility for making available the GenBank DNA seq ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
UniProt
UniProt is a freely accessible database of protein sequence and functional information, many entries being derived from genome sequencing projects. It contains a large amount of information about the biological function of proteins derived from the research literature. It is maintained by the UniProt consortium, which consists of several European bioinformatics organisations and a foundation from Washington, DC, United States. The UniProt consortium The UniProt consortium comprises the European Bioinformatics Institute (EBI), the Swiss Institute of Bioinformatics (SIB), and the Protein Information Resource (PIR). EBI, located at the Wellcome Trust Genome Campus in Hinxton, UK, hosts a large resource of bioinformatics databases and services. SIB, located in Geneva, Switzerland, maintains the ExPASy (Expert Protein Analysis System) servers that are a central resource for proteomics tools and databases. PIR, hosted by the National Biomedical Research Foundation (NBRF) at the Geo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ensembl
Ensembl genome database project is a scientific project at the European Bioinformatics Institute, which provides a centralized resource for geneticists, molecular biologists and other researchers studying the genomes of our own species and other vertebrates and model organisms. Ensembl is one of several well known genome browsers for the retrieval of genomic information. Similar databases and browsers are found at NCBI and the University of California, Santa Cruz (UCSC). History The human genome consists of three billion base pairs, which code for approximately 20,000–25,000 genes. However the genome alone is of little use, unless the locations and relationships of individual genes can be identified. One option is manual annotation, whereby a team of scientists tries to locate genes using experimental data from scientific journals and public databases. However this is a slow, painstaking task. The alternative, known as automated annotation, is to use the power of computers ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Comparative Genomics
Comparative genomics is a field of biological research in which the genomic features of different organisms are compared. The genomic features may include the DNA sequence, genes, gene order, regulatory sequences, and other genomic structural landmarks. In this branch of genomics, whole or large parts of genomes resulting from genome projects are compared to study basic biological similarities and differences as well as evolutionary relationships between organisms. The major principle of comparative genomics is that common features of two organisms will often be encoded within the DNA that is evolutionarily conserved between them. Therefore, comparative genomic approaches start with making some form of alignment of genome sequences and looking for orthologous sequences (sequences that share a common ancestry) in the aligned genomes and checking to what extent those sequences are conserved. Based on these, genome and molecular evolution are inferred and this may in turn be ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Orthologs
Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a speciation event (orthologs), or a duplication event (paralogs), or else a horizontal (or lateral) gene transfer event (xenologs). Homology among DNA, RNA, or proteins is typically inferred from their nucleotide or amino acid sequence similarity. Significant similarity is strong evidence that two sequences are related by evolutionary changes from a common ancestral sequence. Alignments of multiple sequences are used to indicate which regions of each sequence are homologous. Identity, similarity, and conservation The term "percent homology" is often used to mean "sequence similarity”, that is the percentage of identical residues (''percent identity''), or the percentage of residues conserved with similar physicochemical properties ( ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |