|
Nrf2
Nuclear factor erythroid 2-related factor 2 (NRF2), also known as nuclear factor erythroid-derived 2-like 2, is a transcription factor that in humans is encoded by the ''NFE2L2'' gene. NRF2 is a basic leucine zipper (bZIP) protein that may regulate the expression of antioxidant proteins that protect against oxidative damage triggered by injury and inflammation, according to preliminary research. In vitro, NRF2 binds to antioxidant response elements (AREs) in the promoter regions of genes encoding cytoprotective proteins. NRF2 induces the expression of heme oxygenase 1 ''in vitro'' leading to an increase in phase II enzymes. NRF2 also inhibits the NLRP3 inflammasome. NRF2 appears to participate in a complex regulatory network and performs a pleiotropic role in the regulation of metabolism, inflammation, autophagy, proteostasis, mitochondrial physiology, and immune responses. Several drugs that stimulate the NFE2L2 pathway are being studied for treatment of diseases that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
KEAP1
Kelch-like ECH-associated protein 1 is a protein that in humans is encoded by the ''Keap1'' gene. Structure Keap1 has four discrete protein domains. The N-terminal Broad complex, Tramtrack and Bric-à-Brac (BTB) domain contains the Cys151 residue, which is one of the important cysteines in stress sensing. The intervening region (IVR) domain contains two critical cysteine residues, Cys273 and Cys288, which are a second group of cysteines important for stress sensing. A double glycine repeat (DGR) and C-terminal region (CTR) domains collaborate to form a β-propeller structure, which is where Keap1 interacts with Nrf2. Interactions Keap1 has been shown to interact with Nrf2, a master regulator of the antioxidant response, which is important for the amelioration of oxidative stress. Under quiescent conditions, Nrf2 is anchored in the cytoplasm through binding to Keap1, which, in turn, facilitates the ubiquitination and subsequent proteolysis of Nrf2. Such sequestratio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
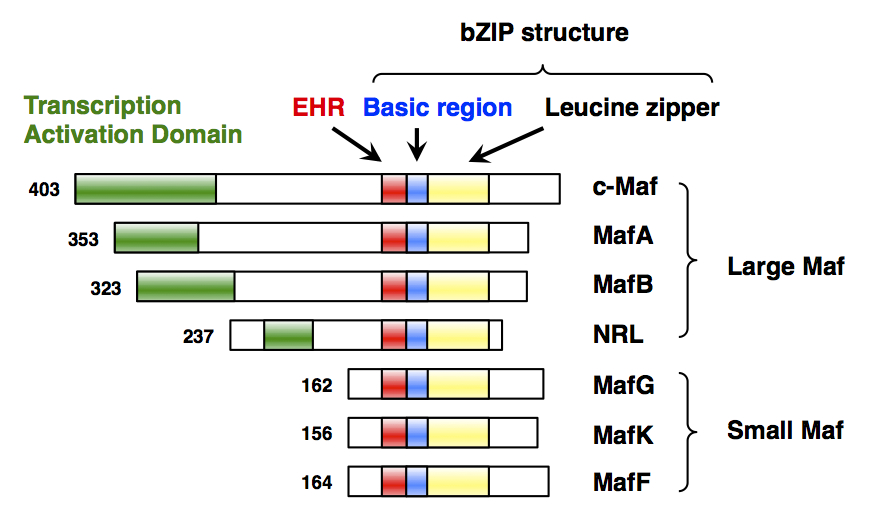
MAFG
Transcription factor MafG is a bZip Maf transcription factor protein that in humans is encoded by the ''MAFG'' gene. MafG is one of the small Maf proteins, which are basic region and leucine zipper (bZIP)-type transcription factors. The HUGO Gene Nomenclature Committee-approved gene name of ''MAFG'' is “v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G”. Discovery MafG was first cloned and identified in chicken in 1995 as a new member of the small Maf (sMaf) genes. MAFG has been identified in many vertebrates, including humans. There are three functionally redundant sMaf proteins in vertebrates, MafF, MafG, and MafK. Structure MafG has a bZIP structure that consists of a basic region for DNA binding and a leucine zipper structure for dimer formation. Similar to other sMafs, MafG lacks any canonical transcriptional activation domains. Expression ''MAFG'' is broadly but differentially expressed in various tissues. ''MAFG'' expression was detected in a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NFE2L1
Nuclear factor erythroid 2-related factor 1 (Nrf1) also known as nuclear factor erythroid-2-like 1 (NFE2L1) is a protein that in humans is encoded by the ''NFE2L1'' gene. Since NFE2L1 is referred to as Nrf1, it is often confused with nuclear respiratory factor 1 (Nrf1). NFE2L1 is a cap ‘n’ collar, basic-leucine zipper (bZIP) transcription factor. Several isoforms of NFE2L1 have been described for both human and mouse genes. NFE2L1 was first cloned in yeast using a genetic screening method. NFE2L1 is ubiquitously expressed, and high levels of transcript are detected in the heart, kidney, skeletal muscle, fat, and brain. Four separate regions — an asparagine/serine/threonine, acidic domains near the N-terminus, and a serine-rich domain located near the CNC motif — are required for full transactivation function of NFE2L1. NFE2L1 is a key regulator of cellular functions including oxidative stress response, differentiation, inflammatory response, metabolism, cholesterol han ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cullin 3
Cullin 3 is a protein that in humans is encoded by the ''CUL3'' gene. Cullin 3 protein belongs to the family of cullins which in mammals contains eight proteins (Cullin 1, Cullin 2, Cullin 3, Cullin 4A, Cullin 4B, Cullin 5, Cullin 7 and Cullin 9). Cullin proteins are an evolutionarily conserved family of proteins throughout yeast, plants and mammals. Protein function Cullin 3 is a component of Cullin- RING E3 ubiquitin ligases complexes (CRLs) which are involved in protein ubiquitylation and represent a part of ubiquitin–proteasome system (UPS). Added ubiquitin moieties to the lysine residue by CRLs then target the protein for the proteasomal degradation. Cullin- RING E3 ubiquitin ligases are involved in many cellular processes responsible for cell cycle regulation, stress response, protein trafficking, signal transduction, DNA replication, transcription, protein quality control, circadian clock and development. Deletion of ''CUL3'' gene in mice causes embryonic lethality. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MAFF (gene)
Transcription factor MafF is a bZip Maf transcription factor protein that in humans is encoded by the ''MAFF'' gene. MafF is one of the small Maf proteins, which are basic region and leucine zipper (bZIP)-type transcription factors. The HUGO Gene Nomenclature Committee-approved gene name of ''MAFF'' is “v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F”. Discovery MafF was first cloned and identified in chicken in 1993 as a member of the small Maf (sMaf) genes. MAFF has been identified in many vertebrates, including humans. There are three functionally redundant sMaf proteins in vertebrates, MafF, MafG, and MafK. Structure MafF has a bZIP structure that consists of a basic region for DNA binding and a leucine zipper structure for dimer formation. Similar to other sMafs, MafF lacks any canonical transcriptional activation domains. Expression ''MAFF'' is broadly but differentially expressed in various tissues. ''MAFF'' expression was detected in all 1 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
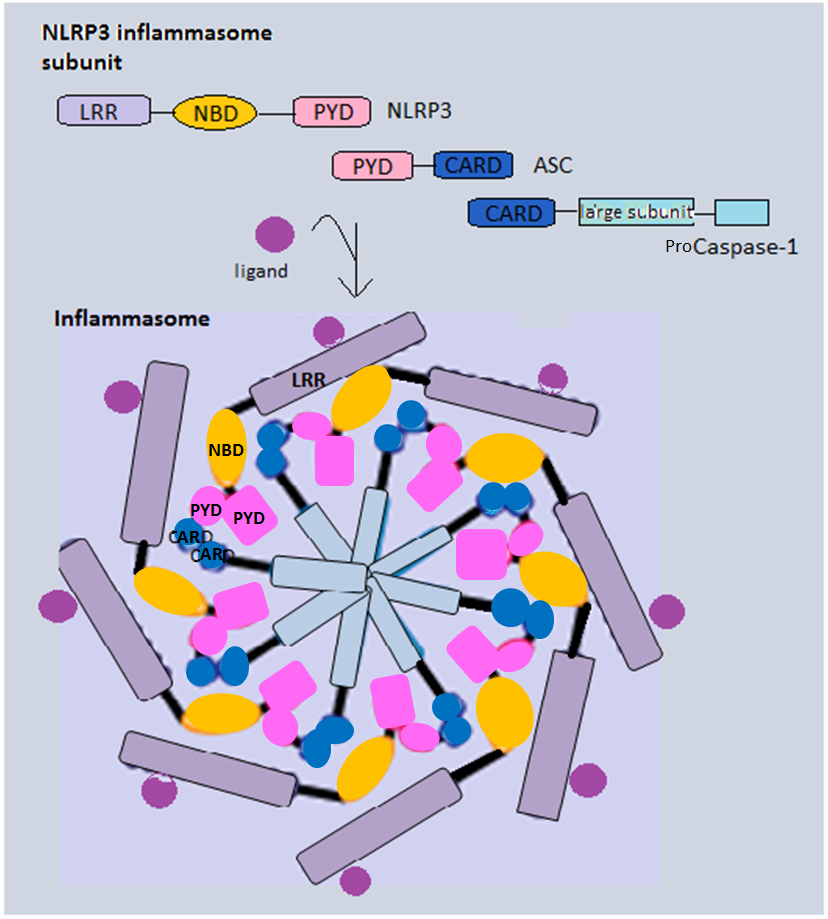
Inflammasome
Inflammasomes are cytosolic multiprotein oligomers of the innate immune system responsible for the activation of inflammatory responses. Activation and assembly of the inflammasome promotes proteolytic cleavage, maturation and secretion of pro-inflammatory cytokines interleukin 1β (IL-1β) and interleukin 18 (IL-18), as well as cleavage of Gasdermin-D. The N-terminal fragment resulting from this cleavage induces a pro-inflammatory form of programmed cell death distinct from apoptosis, referred to as pyroptosis, and is responsible for secretion of the mature cytokines, presumably through the formation of pores in the plasma membrane. Inflammasome activation is initiated by different kinds of cytosolic pattern recognition receptors (PRRs) that respond to either microbe-derived pathogen-associated molecular patterns (PAMPs) or danger-associated molecular patterns (DAMPs) generated by the host cell. Pattern recognition receptors involved in inflammasomes comprise NLRs (nucleotide ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-Fos
Protein c-Fos is a proto-oncogene that is the human homolog of the retroviral oncogene v-fos. It is encoded in humans by the ''FOS'' gene. It was first discovered in rat fibroblasts as the transforming gene of the FBJ MSV (Finkel–Biskis–Jinkins murine osteogenic sarcoma virus) (Curran and Tech, 1982). It is a part of a bigger Fos family of transcription factors which includes c-Fos, FosB, Fra-1 and Fra-2. It has been mapped to chromosome region 14q21→q31. c-Fos encodes a 62 kDa protein, which forms heterodimer with c-jun (part of Jun family of transcription factors), resulting in the formation of AP-1 (Activator Protein-1) complex which binds DNA at AP-1 specific sites at the promoter and enhancer regions of target genes and converts extracellular signals into changes of gene expression. It plays an important role in many cellular functions and has been found to be overexpressed in a variety of cancers. Structure and function c-Fos is a 380 amino acid protein with a bas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-jun
Transcription factor Jun is a protein that in humans is encoded by the ''JUN'' gene. c-Jun, in combination with protein c-Fos, forms the AP-1 early response transcription factor. It was first identified as the Fos-binding protein p39 and only later rediscovered as the product of the JUN gene. c-jun was the first oncogenic transcription factor discovered. The proto-oncogene c-Jun is the cellular homolog of the viral oncoprotein v-jun (). The viral homolog v-jun was discovered in avian sarcoma virus 17 and was named for ''ju-nana'', the Japanese word for 17. The human JUN encodes a protein that is highly similar to the viral protein, which interacts directly with specific target DNA sequences to regulate gene expression. This gene is intronless and is mapped to 1p32-p31, a chromosomal region involved in both translocations and deletions in human malignancies. Function Regulation Both Jun and its dimerization partners in AP-1 formation are subject to regulation by diverse ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NFE2L3
Nuclear factor (erythroid 2)-like factor 3, also known as NFE2L3 or 'NRF3', is a transcription factor that in humans is encoded by the ''Nfe2l3'' gene. Nrf3 is a basic leucine zipper (bZIP) transcription factor belonging to the Cap ‘n’ Collar (CNC) family of proteins. In 1989, the first CNC transcription factor NFE2L2 was identified. Subsequently, several related proteins were identified, including NFE2L1 and NFE2L3, in different organisms such as humans, mice, and zebrafish. These proteins are specifically encoded in the humans by ''Nfe2l1'' and ''Nfe2l3'' genes respectively. Gene The ''Nfe2l3'' gene was mapped to the chromosomal location 7p15-p14 by fluorescence in situ hybridization (FISH). It covers 34.93 kB from base 26191830 to 26226754 on the direct DNA strand with an exon count of 4. The gene is found near the HOXA gene cluster, similar to the clustering of p45 NF-E2, NFE2L1, and NFE2L2 near HOXC, HOXB, and HOXD genes respectively. This implies that all four ge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oxidative Stress
Oxidative stress reflects an imbalance between the systemic manifestation of reactive oxygen species and a biological system's ability to readily detoxify the reactive intermediates or to repair the resulting damage. Disturbances in the normal redox state of cells can cause toxic effects through the production of peroxides and free radicals that damage all components of the cell, including proteins, lipids, and DNA. Oxidative stress from oxidative metabolism causes base damage, as well as strand breaks in DNA. Base damage is mostly indirect and caused by the reactive oxygen species generated, e.g., O2− ( superoxide radical), OH (hydroxyl radical) and H2O2 (hydrogen peroxide). Further, some reactive oxidative species act as cellular messengers in redox signaling. Thus, oxidative stress can cause disruptions in normal mechanisms of cellular signaling. In humans, oxidative stress is thought to be involved in the development of attention deficit hyperactivity disorder, cancer ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heme Oxygenase
Heme oxygenase, or haem oxygenase, (HMOX, commonly abbreviated as HO) is an enzyme that catalyzes the degradation of heme to produce biliverdin, ferrous ion, and carbon monoxide. There are many heme degrading enzymes in nature. In general, only aerobic heme degrading enzymes are referred to as HMOX-like enzymes whereas anaerobic enzymes are typically not affiliated with the HMOX family. Heme oxygenase Heme oxygenase (alternatively spelled using haem or oxidase) catalyzes the degradation of heme to biliverdin/ bilirubin, ferrous ion, and carbon monoxide. The human genome may encode three isoforms of HMOX. The degradation of heme forms three distinct chromogens as seen in healing cycle of a bruise. This reaction can occur in virtually every cell and platelet; the classic example is the healing process of a contusion, which forms different chromogens as it gradually heals: (red) heme to (green) biliverdin to (yellow) bilirubin which is widely known for jaundice. In general, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BZIP Domain
The Basic Leucine Zipper Domain (bZIP domain) is found in many DNA binding eukaryotic proteins. One part of the domain contains a region that mediates sequence specific DNA binding properties and the leucine zipper that is required to hold together (dimerize) two DNA binding regions. The DNA binding region comprises a number of basic amino acids such as arginine and lysine. Proteins containing this domain are transcription factors. bZIP transcription factors bZIP transcription factors are found in all eukaryotes and form one of the largest families of dimerizing TFs. An evolutionary study from 2008 revealed that 4 bZIP genes were encoded by the genome of the most recent common ancestor of all plants. Interactions between bZIP transcription factors are numerous and complex and play important roles in cancer development in epithelial tissues, steroid hormone synthesis by cells of endocrine tissues, factors affecting reproductive functions, and several other phenomena that affe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |