|
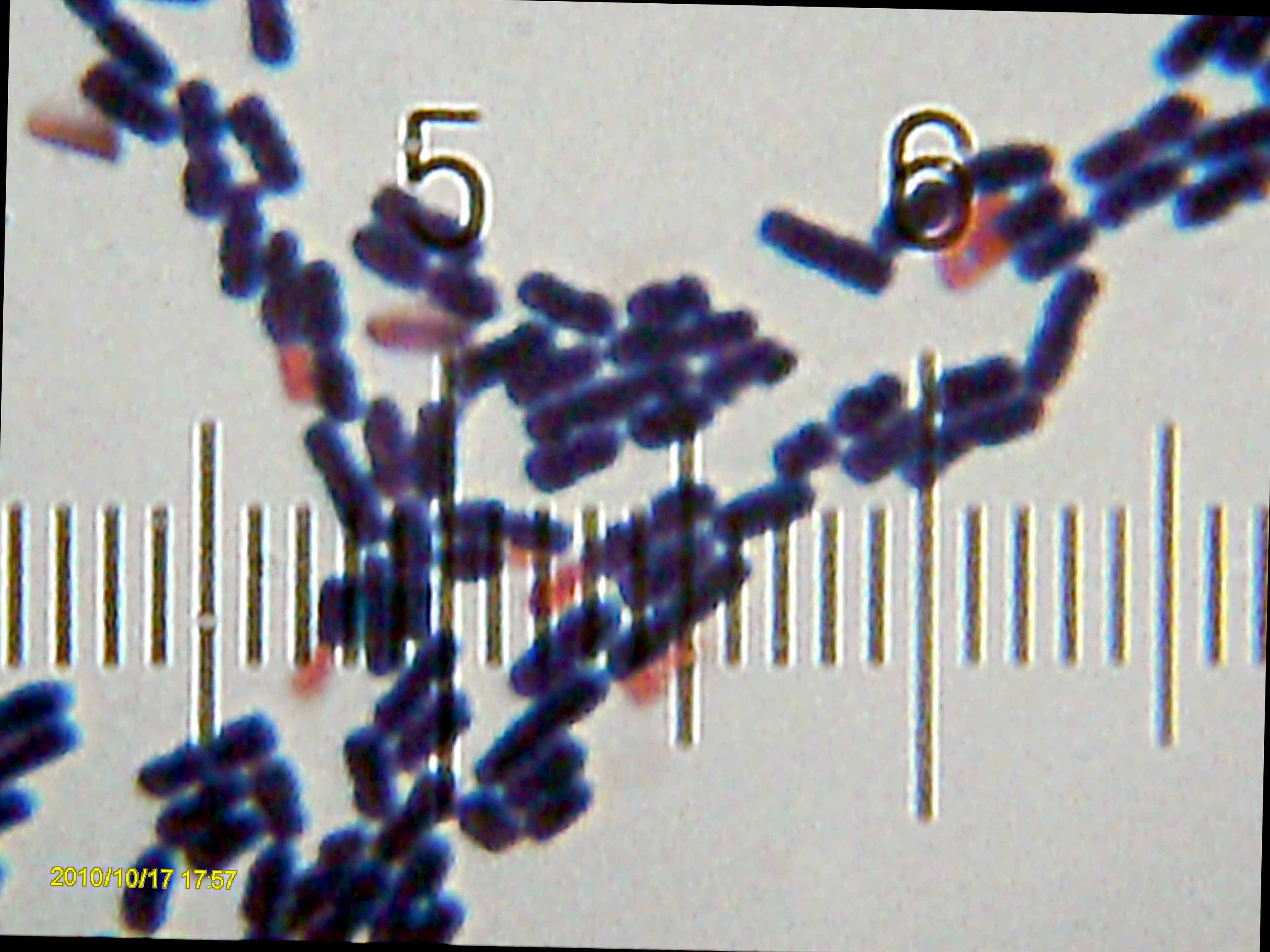
Methanocalculus Chunghsingensis
''Methanocalculus'' is a genus of the Methanomicrobiales, and is known to include methanogens. The genome of ''Methanocalculus'' is somewhat different from other genera of methanogenic archaea, with less than 90% 16S ribosomal RNA similarity. The species within ''Methanocalculus'' also have a greater tolerance to salt than other microorganisms, and they can live at salt concentrations as high as 125 g/L. Some species within ''Methanocalculus'' are neutrophiles, and '' Methanocalculus natronophilus'', discovered in 2013, is a strict alkaliphile. Nomenclature The name "Methanocalculus" has Latin roots: "methano" for methane and "calculus" for gravel. Overall, it means ''gravel-shaped organism that produces methane.'' Phylogeny The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN) and National Center for Biotechnology Information (NCBI). See also * List of Archaea genera This article lists the genera of the Archaea. The ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Archaea
Archaea ( ; singular archaeon ) is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria (in the Archaebacteria kingdom), but this term has fallen out of use. Archaeal cells have unique properties separating them from the other two domains, Bacteria and Eukaryota. Archaea are further divided into multiple recognized phyla. Classification is difficult because most have not been isolated in a laboratory and have been detected only by their gene sequences in environmental samples. Archaea and bacteria are generally similar in size and shape, although a few archaea have very different shapes, such as the flat, square cells of ''Haloquadratum walsbyi''. Despite this morphological similarity to bacteria, archaea possess genes and several metabolic pathways that are more closely related to those of eukaryotes, notably for the enzymes involved ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
16S Ribosomal RNA
16 S ribosomal RNA (or 16 S rRNA) is the RNA component of the 30S subunit of a prokaryotic ribosome (SSU rRNA). It binds to the Shine-Dalgarno sequence and provides most of the SSU structure. The genes coding for it are referred to as 16S rRNA gene and are used in reconstructing phylogenies, due to the slow rates of evolution of this region of the gene. Carl Woese and George E. Fox were two of the people who pioneered the use of 16S rRNA in phylogenetics in 1977. Multiple sequences of the 16S rRNA gene can exist within a single bacterium. Functions * Like the large (23S) ribosomal RNA, it has a structural role, acting as a scaffold defining the positions of the ribosomal proteins. * The 3-end contains the anti- Shine-Dalgarno sequence, which binds upstream to the AUG start codon on the mRNA. The 3-end of 16S RNA binds to the proteins S1 and S21 which are known to be involved in initiation of protein synthesis * Interacts with 23S, aiding in the binding of the two ribosomal s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Archaea Genera
This article lists the genera of the Archaea. The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN) and National Center for Biotechnology Information (NCBI). Phylogeny National Center for Biotechnology Information (NCBI) taxonomy was initially used to decorate the genome tree via tax2tree. The 16S rRNA-based Greengenes taxonomy is used to supplement the taxonomy particularly in regions of the tree with no cultured representatives. List of Prokaryotic names with Standing in Nomenclature (LPSN) is used as the primary taxonomic authority for establishing naming priorities. Taxonomic ranks are normalised using phylorank and the taxonomy manually curated to remove polyphyletic groups. Cladogram was taken from the GTDB release 07-RS207 (8th April 2022). The position of clades with a "question mark" are based on the additional phylogeny of the 16S rRNA-based LTP_12_2021 by The All-Species Living Tree Project. Phylum " Altarcha ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methanocorpusculum
In taxonomy, ''Methanocorpusculum'' is a genus of microbes within the family Methanocorpusculaceae. The species within ''Methanocorpusculum'' were first isolated from biodisgester wastewater and activated sludge from anaerobic digestors. In nature, they live in freshwater environments. Unlike most other methanogenic archaea, they do not require high temperatures or extreme salt concentrations to live and grow. Nomenclature The name ''Methanocorpusculum'' has Latin roots. It means ''bodies that produce methane''. Description and metabolism The cells of these archaea are small, irregular, and coccoid in shape. They are Gram-negatives y and not very motile. They reduce carbon dioxide to methane using hydrogen, but they can also use formate or secondary alcohols. They cannot use acetate or methylamines. They grow fastest at temperatures of 30–40 °C. Phylogeny The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature ( ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome Taxonomy Database
The Genome Taxonomy Database (GTDB) is an online database that maintains information on a proposed nomenclature of prokaryotes, following a phylogenomic approach based on a set of conserved single-copy proteins. In addition to breaking up paraphyletic groups, this method also reassigns taxonomic ranks algorithmically, creating new names in both cases. Information for archaea was added in 2020, along with a species classification based on average nucleotide identity. Each update incorporates new genomes as well as human adjustments to the taxonomy. An open-source tool called GTDB-Tk is available to classify draft genomes into the GTDB hierarchy. The GTDB system, via GTDB-Tk, has been used to catalogue not-yet-named bacteria in the human gut microbiome and other metagenomic sources. The GTDB is incorporated into the ''Bergey's Manual of Systematics of Archaea and Bacteria'' in 2019 as its phylogenomic resource. See also * PhyloCode * National Center for Biotechnology Informa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
The All-Species Living Tree Project
The All-Species Living Tree' Project is a collaboration between various academic groups/institutes, such as ARB Project, ARB, SILVA rRNA database project, and List of Prokaryotic names with Standing in Nomenclature, LPSN, with the aim of assembling a database of 16S ribosomal RNA, 16S rRNA sequences of all validly published species of ''Bacteria'' and ''Archaea''. At one stage, 23S ribosomal RNA, 23S sequences were also collected, but this has since stopped. Currently there are over 10,950 species in the aligned dataset and several more are being added either as new species are discovered or species that are not represented in the database are sequenced. Initially the latter group consisted of 7% of species. Similar (and more recent) projects include the Genomic Encyclopedia of Bacteria and Archaea (GEBA), which focused on whole genome sequencing of bacteria and archaea. Tree The tree was created by maximum likelihood analysis without bootstrap: consequently accuracy is traded ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
National Center For Biotechnology Information
The National Center for Biotechnology Information (NCBI) is part of the United States National Library of Medicine (NLM), a branch of the National Institutes of Health (NIH). It is approved and funded by the government of the United States. The NCBI is located in Bethesda, Maryland, and was founded in 1988 through legislation sponsored by US Congressman Claude Pepper. The NCBI houses a series of databases relevant to biotechnology and biomedicine and is an important resource for bioinformatics tools and services. Major databases include GenBank for DNA sequences and PubMed, a bibliographic database for biomedical literature. Other databases include the NCBI Epigenomics database. All these databases are available online through the Entrez search engine. NCBI was directed by David Lipman, one of the original authors of the BLAST sequence alignment program and a widely respected figure in bioinformatics. GenBank NCBI had responsibility for making available the GenBank DNA seque ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Prokaryotic Names With Standing In Nomenclature
List of Prokaryotic names with Standing in Nomenclature (LPSN) is an online database that maintains information on the naming and taxonomy of prokaryotes, following the taxonomy requirements and rulings of the International Code of Nomenclature of Prokaryotes The International Code of Nomenclature of Prokaryotes (ICNP) formerly the International Code of Nomenclature of Bacteria (ICNB) or Bacteriological Code (BC) governs the scientific names for Bacteria and Archaea.P. H. A. Sneath, 2003. A short hist .... The database was curated from 1997 to June 2013 by Jean P. Euzéby. From July 2013 to January 2020, LPSN was curated by Aidan C. Parte. In February 2020, a new version of LPSN was published as a service of the Leibniz Institute DSMZ, thereby also integrating the Prokaryotic Nomenclature Up-to-date service. References External links List of Prokaryotic names with Standing in Nomenclature [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methane
Methane ( , ) is a chemical compound with the chemical formula (one carbon atom bonded to four hydrogen atoms). It is a group-14 hydride, the simplest alkane, and the main constituent of natural gas. The relative abundance of methane on Earth makes it an economically attractive fuel, although capturing and storing it poses technical challenges due to its gaseous state under normal conditions for temperature and pressure. Naturally occurring methane is found both below ground and under the seafloor and is formed by both geological and biological processes. The largest reservoir of methane is under the seafloor in the form of methane clathrates. When methane reaches the surface and the atmosphere, it is known as atmospheric methane. The Earth's atmospheric methane concentration has increased by about 150% since 1750, and it accounts for 20% of the total radiative forcing from all of the long-lived and globally mixed greenhouse gases. It has also been detected on other plane ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alkaliphile
Alkaliphiles are a class of extremophilic microbes capable of survival in alkaline ( pH roughly 8.5–11) environments, growing optimally around a pH of 10. These bacteria can be further categorized as obligate alkaliphiles (those that require high pH to survive), facultative alkaliphiles (those able to survive in high pH, but also grow under normal conditions) and haloalkaliphiles (those that require high salt content to survive).HORIKOSHI, KOKI. "Alkaliphiles: Some Applications of Their Products for Biotechnology." MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS 63.4 (1999): 735-50. Print. Background information Microbial growth in alkaline conditions presents several complications to normal biochemical activity and reproduction, as high pH is detrimental to normal cellular processes. For example, alkalinity can lead to denaturation of DNA, instability of the plasma membrane and inactivation of cytosolic enzymes, as well as other unfavorable physiological changes.Higashibata, Akira, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Neutrophile
A neutrophile is a neutrophilic organism that thrives in a neutral pH environment between 6.5 and 7.5. Environment The pH of the environment can support growth or hinder neutrophilic organisms. When the pH is within the microbe's range, they grow and within that range there is an optimal growth pH. Neutrophiles are adapted to live in an environment where the hydrogen ion concentration is at equilibrium. They are sensitive to the concentration, and when the pH become too basic or acidic, the cell's proteins can denature. Depending on the microbe and the pH, the microbe's growth can be slowed or stopped altogether. Manipulation of the pH of the environment that the microbe is in is used by the food industry to control its growth in order to increase the shelf life of food. See also * Acidophile * Acidophobe * Alkaliphile * Extremophile * Mesophile A mesophile is an organism that grows best in moderate temperature, neither too hot nor too cold, with an optimum growth range f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
.jpg)