|
Lysosome-associated Membrane Glycoproteins
Lysosome-associated membrane glycoproteins (LAMPs) are integral membrane proteins, specific to lysosomes, and whose exact biological function is not yet clear. Structurally, the lamp proteins consist of two internally homologous lysosome-luminal domains separated by a proline-rich hinge region; at the C-terminal extremity there is a transmembrane region (TM) followed by a very short cytoplasmic tail (C). In each of the duplicated domains, there are two conserved disulfide bonds. This structure is schematically represented in the figure below. +-----+ +-----+ +-----+ +-----+ , , , , , , , , +--------------------------++Hinge++--------------------------++TM++C+ In mammals, there are two closely related types of lamp: LAMP1 and LAMP2. CD69 (also called gp110 or macrosialin) is a heavily glycosylated integral membrane protein whose structure consists of a mucin-like domain followed by a pro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
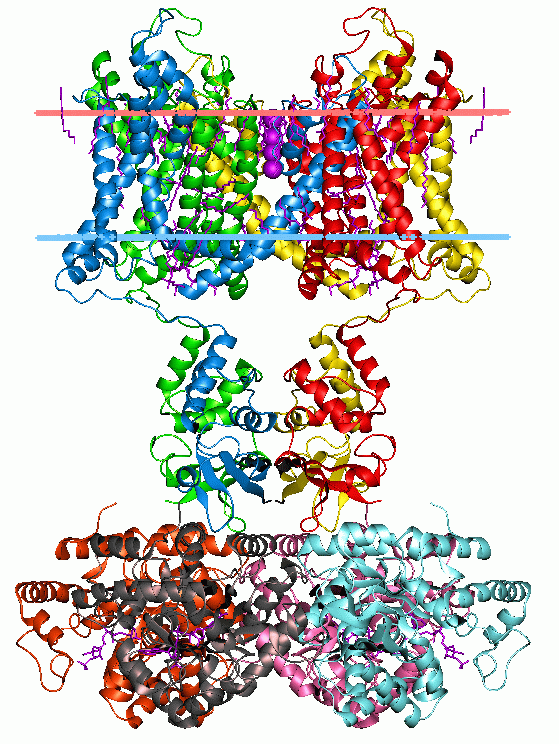
Membrane Protein
Membrane proteins are common proteins that are part of, or interact with, biological membranes. Membrane proteins fall into several broad categories depending on their location. Integral membrane proteins are a permanent part of a cell membrane and can either penetrate the membrane (transmembrane) or associate with one or the other side of a membrane ( integral monotopic). Peripheral membrane proteins are transiently associated with the cell membrane. Membrane proteins are common, and medically important—about a third of all human proteins are membrane proteins, and these are targets for more than half of all drugs. Nonetheless, compared to other classes of proteins, determining membrane protein structures remains a challenge in large part due to the difficulty in establishing experimental conditions that can preserve the correct conformation of the protein in isolation from its native environment. Function Membrane proteins perform a variety of functions vital to the sur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lysosome
A lysosome () is a membrane-bound organelle found in many animal cells. They are spherical vesicles that contain hydrolytic enzymes that can break down many kinds of biomolecules. A lysosome has a specific composition, of both its membrane proteins, and its lumenal proteins. The lumen's pH (~4.5–5.0) is optimal for the enzymes involved in hydrolysis, analogous to the activity of the stomach. Besides degradation of polymers, the lysosome is involved in various cell processes, including secretion, plasma membrane repair, apoptosis, cell signaling, and energy metabolism. Lysosomes act as the waste disposal system of the cell by digesting used materials in the cytoplasm, from both inside and outside the cell. Material from outside the cell is taken up through endocytosis, while material from the inside of the cell is digested through autophagy. The sizes of the organelles vary greatly—the larger ones can be more than 10 times the size of the smaller ones. They were discov ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Disulfide Bond
In biochemistry, a disulfide (or disulphide in British English) refers to a functional group with the structure . The linkage is also called an SS-bond or sometimes a disulfide bridge and is usually derived by the coupling of two thiol groups. In biology, disulfide bridges formed between thiol groups in two cysteine residues are an important component of the secondary and tertiary structure of proteins. ''Persulfide'' usually refers to compounds. In inorganic chemistry disulfide usually refers to the corresponding anion (−S−S−). Organic disulfides Symmetrical disulfides are compounds of the formula . Most disulfides encountered in organo sulfur chemistry are symmetrical disulfides. Unsymmetrical disulfides (also called heterodisulfides) are compounds of the formula . They are less common in organic chemistry, but most disulfides in nature are unsymmetrical. Properties The disulfide bonds are strong, with a typical bond dissociation energy of 60 kcal/mol (251& ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
LAMP1
Lysosomal-associated membrane protein 1 (LAMP-1) also known as lysosome-associated membrane glycoprotein 1 and CD107a (Cluster of Differentiation 107a), is a protein that in humans is encoded by the ''LAMP1'' gene. The human ''LAMP1'' gene is located on the long arm (q) of chromosome 13 at region 3, band 4 (13q34). Lysosomal-associated membrane protein 1 is a glycoprotein from a family of Lysosome-associated membrane glycoproteins. The LAMP-1 glycoprotein is a type I transmembrane protein which is expressed at high or medium levels in at least 76 different normal tissue cell types. It resides primarily across l ysosomal membranes, and functions to provide selectins with carbohydrate ligands. CD107a has also been shown to be a marker of degranulation on lymphocytes such as CD8+ and NK cells, and may also play a role in tumor cell differentiation and metastasis. Structure Residing primarily across lysosomal membranes, these glycoproteins consist of a large, highly glycosylated e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
LAMP2
Lysosome-associated membrane protein 2 (LAMP2), also known as CD107b (Cluster of Differentiation 107b) and Mac-3, is a human gene. Its protein, LAMP2, is one of the lysosome-associated membrane glycoproteins. The protein encoded by this gene is a member of a family of membrane glycoproteins. This glycoprotein provides selectins with carbohydrate ligands. It may play a role in tumor cell metastasis. It may also function in the protection, maintenance, and adhesion of the lysosome. Alternative splicing of the gene produces three variants - LAMP-2A, LAMP-2B and LAMP-2C. LAMP-2A is the receptor for chaperone-mediated autophagy. Recently it has been determined that antibodies against LAMP-2 account for a fraction of patients who get a serious kidney disease termed focal necrotizing glomerulonephritis. LAMP-2B is associated with Danon disease. Structure and tissue distribution The gene for LAMP2 has 9 coding exons and 2 alternate last exons, 9a and 9b. When the last exon is spli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CD69
CD69 (Cluster of Differentiation 69) is a human transmembrane C-Type lectin protein encoded by the gene. It is an early activation marker that is expressed in hematopoietic stem cells, T cells, and many other cell types in the immune system. It is also implicated in T cell differentiation as well as lymphocyte retention in lymphoid organs. Function The activation of T lymphocytes and Natural Killer (NK) cells, both in vivo and in vitro, induces expression of CD69. This molecule, which appears to be the earliest inducible cell surface glycoprotein acquired during lymphoid activation, is involved in lymphocyte proliferation and functions as a signal-transmitting receptor in lymphocytes, including natural killer (NK) cells, and platelets (Cambiaggi et al., 1992) upplied by OMIM Structure and ligands The gene encoding CD69 is located in the NK gene complex on chromosome 6 and chromosome 12 in mice and humans respectively. Activation signaling pathways in lymphocytes, NK cells, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CD68
CD68 ( Cluster of Differentiation 68) is a protein highly expressed by cells in the monocyte lineage (e.g., monocytic phagocytes, osteoclasts), by circulating macrophages, and by tissue macrophages (e.g., Kupffer cells, microglia). Structure and function Human CD68 is a Type I transmembrane glycoprotein, heavily glycosylated in its extracellular domain, with a molecular weight of 110 kD. Its primary sequence consists of 354 amino acids with predicted molecular weight of 37.4 kD if it were not glycosylated. The human CD68 protein is encoded by the "CD68" gene which maps to Chromosome 17. Other names or aliases for this gene in humans and other animals include: CD68 Molecule, CD68 Antigen, GP110, Macrosialin, Scavenger Receptor Class D, Member 1, SCARD1, and LAMP4. The mouse equivalent is known as "macrosialin". CD68 is functionally and evolutionarily related to other gene/protein family members, including: * the hematopoietic mucin-like family of molecules that includes leuko ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
LAMP3
Lysosome-associated membrane glycoprotein 3 (LAMP3, Lamp3) is a protein that in humans is encoded by the ''LAMP3'' gene. It is one of the lysosome-associated membrane glycoproteins. LAMP3 also known as DC-LAMP (Dendritic cell lysosomal associated membrane glycoprotein) is a member of the LAMP family along with LAMP1 and LAMP2, these proteins make up the members of the glycoconjugate coat present on the inside of the lysosomal membrane. In humans, this protein is almost exclusively found in mature Dendritic cells. While LAMP3 can be observed on the surface of dendritic cells, the protein is mainly found within lysosomes. LAMP3 first appears in the MHC Class II compartment and in cells aids in the identifying and processing of an antigen during an immune response. LAMP3 protein is linked with the maturation of dendritic cells, and as a marker for transformed type II pneumocytes or alveolar cells. Studies have linked LAMP3 with the inhibition of the viral replication of Influenza A ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Domains
In molecular biology, a protein domain is a region of a protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins. Background The concept of the domain was first proposed in 1973 by Wetlaufer after ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Families
A protein family is a group of evolutionarily related proteins. In many cases, a protein family has a corresponding gene family, in which each gene encodes a corresponding protein with a 1:1 relationship. The term "protein family" should not be confused with family as it is used in taxonomy. Proteins in a family descend from a common ancestor and typically have similar three-dimensional structures, functions, and significant sequence similarity. The most important of these is sequence similarity (usually amino-acid sequence), since it is the strictest indicator of homology and therefore the clearest indicator of common ancestry. A fairly well developed framework exists for evaluating the significance of similarity between a group of sequences using sequence alignment methods. Proteins that do not share a common ancestor are very unlikely to show statistically significant sequence similarity, making sequence alignment a powerful tool for identifying the members of protein familie ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |