|
List Of Alignment Visualization Software
This page is a subsection of the list of sequence alignment software. Multiple alignment visualization tools typically serve four purposes: * Aid general understanding of large-scale DNA or protein alignments * Visualize alignments for figures and publication * Manually edit and curate automatically generated alignments * Analysis in depth The rest of this article is focused on only multiple global alignments of homologous proteins. The first two are a natural consequence of most representations of alignments and their annotation being human-unreadable and best portrayed in the familiar sequence row and alignment column format, of which examples are widespread in the literature. The third is necessary because algorithms for both ''multiple sequence alignment'' and ''structural alignment'' use heuristics which do not always perform perfectly. The fourth is a great example of how interactive graphical tools enable a worker involved in sequence analysis to conveniently execute a varie ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Sequence Alignment Software
This list of sequence alignment software is a compilation of software tools and web portals used in pairwise sequence alignment and multiple sequence alignment. See structural alignment software for structural alignment of proteins. Database search only *Sequence type: protein or nucleotide Pairwise alignment *Sequence type: protein or nucleotide **Alignment type: local or global Multiple sequence alignment *Sequence type: protein or nucleotide. **Alignment type: local or global Genomics analysis *Sequence type: protein or nucleotide Motif finding *Sequence type: protein or nucleotide Benchmarking Alignment viewers, editors Please see List of alignment visualization software. Short-read sequence alignment See also * List of open source bioinformatics software References {{Reflist Genetics-related lists, Sequence Lists of bioinformatics software, Sequence alignment software ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Neighbor Joining
In bioinformatics, neighbor joining is a bottom-up (agglomerative) clustering method for the creation of phylogenetic trees, created by Naruya Saitou and Masatoshi Nei in 1987. Usually based on DNA or protein sequence data, the algorithm requires knowledge of the distance between each pair of taxa (e.g., species or sequences) to create the phylogenetic tree. The algorithm Neighbor joining takes a distance matrix, which specifies the distance between each pair of taxa, as input. The algorithm starts with a completely unresolved tree, whose topology corresponds to that of a star network, and iterates over the following steps, until the tree is completely resolved, and all branch lengths are known: # Based on the current distance matrix, calculate a matrix Q (defined below). # Find the pair of distinct taxa i and j (i.e. with i \neq j) for which Q(i,j) is smallest. Make a new node that joins the taxa i and j, and connect the new node to the central node. For example, in part (B ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
XSLT
XSLT (Extensible Stylesheet Language Transformations) is a language originally designed for transforming XML documents into other XML documents, or other formats such as HTML for web pages, plain text or XSL Formatting Objects, which may subsequently be converted to other formats, such as PDF, PostScript and PNG. Support for JSON and plain-text transformation was added in later updates to the XSLT 1.0 specification. As of August 2022, the most recent stable version of the language is XSLT 3.0, which achieved Recommendation status in June 2017. XSLT 3.0 implementations support Java, .NET, C/C++, Python, PHP and NodeJS. An XSLT 3.0 Javascript library can also be hosted within the Web Browser. Modern web browsers also include native support for XSLT 1.0. For an XSLT document transformation, the original document is not changed; rather, a new document is created based on the content of an existing one. Typically, input documents are XML files, but anything from which the proce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MIT License
The MIT License is a permissive free software license originating at the Massachusetts Institute of Technology (MIT) in the late 1980s. As a permissive license, it puts only very limited restriction on reuse and has, therefore, high license compatibility. Unlike copyleft software licenses, the MIT License also permits reuse within proprietary software, provided that all copies of the software or its substantial portions include a copy of the terms of the MIT License and also a copyright notice. , the MIT License was the most popular software license found in one analysis, continuing from reports in 2015 that the MIT License was the most popular software license on GitHub. Notable projects that use the MIT License include the X Window System, Ruby on Rails, Nim, Node.js, Lua, and jQuery. Notable companies using the MIT License include Microsoft ( .NET), Google ( Angular), and Meta ( React). License terms The MIT License has the identifier MIT in the SPDX License List. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Information Resource
The Protein Information Resource (PIR), located at Georgetown University Medical Center, is an integrated public bioinformatics resource to support genomic and proteomic research, and scientific studies. It contains protein sequences databases History PIR was established in 1984 by the National Biomedical Research Foundation as a resource to assist researchers and customers in the identification and interpretation of protein sequence information. Prior to that, the foundation compiled the first comprehensive collection of macromolecular sequences in the Atlas of Protein Sequence and Structure, published from 1964 to 1974 under the editorship of Margaret Dayhoff. Dayhoff and her research group pioneered in the development of computer methods for the comparison of protein sequences, for the detection of distantly related sequences and duplications within sequences, and for the inference of evolutionary histories from alignments of protein sequences. Winona Barker and Robert Ledl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Public Domain
The public domain (PD) consists of all the creative work to which no exclusive intellectual property rights apply. Those rights may have expired, been forfeited, expressly waived, or may be inapplicable. Because those rights have expired, anyone can legally use or reference those works without permission. As examples, the works of William Shakespeare, Ludwig van Beethoven, Leonardo da Vinci and Georges Méliès are in the public domain either by virtue of their having been created before copyright existed, or by their copyright term having expired. Some works are not covered by a country's copyright laws, and are therefore in the public domain; for example, in the United States, items excluded from copyright include the formulae of Newtonian physics, cooking recipes,Copyright Protection No ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
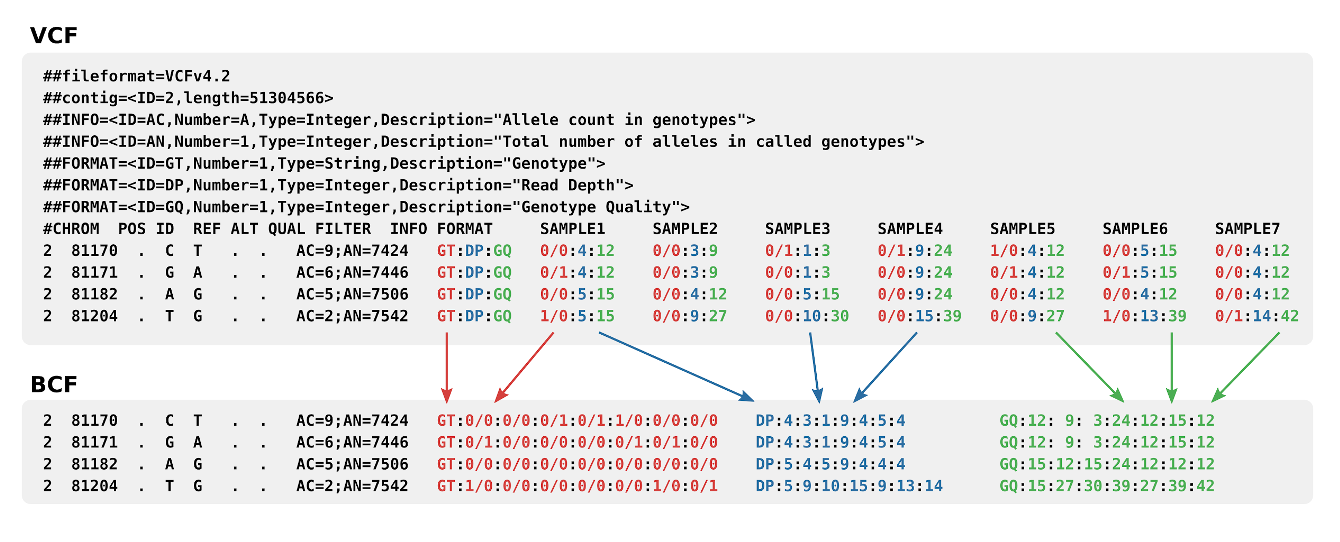
Variant Call Format
The Variant Call Format (VCF) specifies the format of a text file used in bioinformatics for storing gene sequence variations. The format has been developed with the advent of large-scale genotyping and DNA sequencing projects, such as the 1000 Genomes Project. Existing formats for genetic data such as General feature format (GFF) stored all of the genetic data, much of which is redundant because it will be shared across the genomes. By using the variant call format only the variations need to be stored along with a reference genome. The standard is currently in version 4.3, although the 1000 Genomes Project has developed its own specification for structural variations such as duplications, which are not easily accommodated into the existing schema. There is also a genomic VCF (gVCF) extended format, which includes additional information about "blocks" that match the reference and their qualities. A set of tools is also available for editing and manipulating the files. Exampl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SAM (file Format)
Sequence Alignment Map (SAM) is a text-based format originally for storing biological sequences aligned to a reference sequence developed by Heng Li and Bob Handsaker ''et al''. It was developed when the 1000 Genomes Project wanted to move away from the MAQ mapper format and decided to design a new format. The overall TAB-delimited flavour of the format came from an earlier format inspired by BLAT’s PSL. The name of SAM came from Gabor Marth from University of Utah, who originally had a format under the same name but with a different syntax more similar to a BLAST output. It is widely used for storing data, such as nucleotide sequences, generated by next generation sequencing technologies, and the standard has been broadened to include unmapped sequences. The format supports short and long reads (up to 128 Mbp) produced by different sequencing platforms and is used to hold mapped data within the Genome Analysis Toolkit (GATK) and across the Broad Institute, the Wellcome ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FASTA
FASTA is a DNA and protein sequence alignment software package first described by David J. Lipman and William R. Pearson in 1985. Its legacy is the FASTA format which is now ubiquitous in bioinformatics. History The original FASTA program was designed for protein sequence similarity searching. Because of the exponentially expanding genetic information and the limited speed and memory of computers in the 1980s heuristic methods were introduced aligning a query sequence to entire data-bases. FASTA, published in 1987, added the ability to do DNA:DNA searches, translated protein:DNA searches, and also provided a more sophisticated shuffling program for evaluating statistical significance. There are several programs in this package that allow the alignment of protein sequences and DNA sequences. Nowadays, increased computer performance makes it possible to perform searches for local alignment detection in a database using the Smith–Waterman algorithm. FASTA is pronounced "f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sublime Text
Sublime Text is a shareware text and source code editor available for Windows, macOS, and Linux. It natively supports many programming languages and markup languages. Users can customize it with themes and expand its functionality with plugins Plug-in, plug in or plugin may refer to: * Plug-in (computing) is a software component that adds a specific feature to an existing computer program. ** Audio plug-in, adds audio signal processing features ** Photoshop plugin, a piece of software t ..., typically community-built and maintained under free-software licenses. To facilitate plugins, Sublime Text features a Python API. The editor utilizes minimal interface and contains features for programmers including configurable syntax highlighting, code folding, search-and-replace supporting regular-expressions, terminal output window, and more. It is proprietary software, but a free evaluation version is available. Features The following is a list of features of Sublime Te ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gedit
gedit ( or ) is a text editor designed for the GNOME desktop environment. It was GNOME's default text editor and part of the GNOME Core Applications until GNOME version 42 in March 2022, which changed the default text editor to GNOME Text Editor. Designed as a general-purpose text editor, gedit emphasizes simplicity and ease of use, with a clean and simple GUI, according to the philosophy of the GNOME project. It includes tools for editing source code and structured text such as markup languages. It is free and open-source software under the GNU General Public License version 2 or later. gedit is also available for macOS and at one time had version for Windows, but as of May 2020 was no longer available. By July 2017, gedit was not being maintained by any developers, but in August 2017 two developers volunteered to commence work on it again. Features gedit includes syntax highlighting via GtkSourceView for various program code and text markup formats including M ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Less (Unix)
less is a terminal pager program on Unix, Windows, and Unix-like systems used to view (but not change) the contents of a text file one screen at a time. It is similar to , but has the extended capability of allowing both forward and backward navigation through the file. Unlike most Unix text editors/viewers, does not need to read the entire file before starting, allowing for immediate viewing regardless of file size. History Mark Nudelman initially wrote less during 1983–85, in the need of a version of more able to do backward scrolling of the displayed text. The name came from the joke of doing "backwards more." Originally, less was developed for Unix, but it has been ported to a number of other operating systems, including MS-DOS, Microsoft Windows, OS/2, and OS-9, as well as Unix-like systems such as Linux. It is still maintained today by Nudelman. To help remember the difference between less and more, a common joke is to say, "," implying that less has greater function ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |