|
Invertin
Invertase is an enzyme that catalyzes the hydrolysis (breakdown) of sucrose (table sugar) into fructose and glucose. Alternative names for invertase include , saccharase, glucosucrase, beta-h-fructosidase, beta-fructosidase, invertin, sucrase, maxinvert L 1000, fructosylinvertase, alkaline invertase, acid invertase, and the systematic name: beta-fructofuranosidase. The resulting mixture of fructose and glucose is called inverted sugar syrup. Related to invertases are sucrases. Invertases and sucrases hydrolyze sucrose to give the same mixture of glucose and fructose. Invertase is a glycoprotein that hydrolyses (cleaves) the non-reducing terminal beta-fructofuranoside residues. Thus, its systematic name is beta-fructofuranosidase. Invertases cleave the O-C(fructose) bond, whereas the sucrases cleave the O-C(glucose) bond. Invertase cleaves the alpha-1,2-glycosidic bond of sucrose. For industrial use, invertase is usually derived from yeast. It is also synthesized by bees, which use ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Penicillium Chrysogenum
''Penicillium chrysogenum'' (formerly known as ''Penicillium notatum'') is a species of fungus in the genus ''Penicillium''. It is common in temperate and subtropical regions and can be found on salted food products, but it is mostly found in indoor environments, especially in damp or water-damaged buildings. It has been recognised as a species complex that includes ''P. notatum'', ''P. meleagrinum,'' and ''P. cyaneofulvum,'' but molecular phylogeny established that it is a distinct species and that ''P. notatum'' (its popular synonym) is '' P. rubens.'' It has rarely been reported as a cause of human disease. It is the source of several β-lactam antibiotics, most significantly penicillin. Other secondary metabolites of ''P. chrysogenum'' include roquefortine C, meleagrin, chrysogine, 6-MSA YWA1/melanin, andrastatin A, fungisporin, secalonic acids, sorbicillin, and PR-toxin. Like the many other species of the genus ''Penicillium'', ''P. chrysogenum'' usually repro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Enzymes
This article lists enzymes by their classification in the International Union of Biochemistry and Molecular Biology's Enzyme Commission (EC) numbering system. * List of EC numbers (EC 5) * List of EC numbers (EC 6) :Oxidoreductases (EC 1) (Oxidoreductase) *Dehydrogenase * Luciferase *DMSO reductase :EC 1.1 (act on the CH-OH group of donors) * :EC 1.1.1 (with NAD+ or NADP+ as acceptor) ** Alcohol dehydrogenase (NAD) ** Alcohol dehydrogenase (NADP) **Homoserine dehydrogenase ** Aminopropanol oxidoreductase **Diacetyl reductase **Glycerol dehydrogenase **Propanediol-phosphate dehydrogenase ** glycerol-3-phosphate dehydrogenase (NAD+) ** D-xylulose reductase **L-xylulose reductase **Lactate dehydrogenase **Malate dehydrogenase **Isocitrate dehydrogenase ** HMG-CoA reductase * :EC 1.1.2 (with a cytochrome as acceptor) * :EC 1.1.3 (with oxygen as acceptor) **Glucose oxidase **L-gulonolactone oxidase **Thiamine oxidase **Xanthine oxidase * :EC 1.1.4 (with a disul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Tertiary Structure
Protein tertiary structure is the three dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains may interact and bond in a number of ways. The interactions and bonds of side chains within a particular protein determine its tertiary structure. The protein tertiary structure is defined by its atomic coordinates. These coordinates may refer either to a protein domain or to the entire tertiary structure.Branden C. and Tooze J. "Introduction to Protein Structure" Garland Publishing, New York. 1990 and 1991. A number of tertiary structures may fold into a quaternary structure.Kyte, J. "Structure in Protein Chemistry." Garland Publishing, New York. 1995. History The science of the tertiary structure of proteins has progressed from one of hypothesis to one of detailed definition. Although Emil Fischer had suggested proteins were made of polypeptid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrogen Bond
In chemistry, a hydrogen bond (or H-bond) is a primarily electrostatic force of attraction between a hydrogen (H) atom which is covalently bound to a more electronegative "donor" atom or group (Dn), and another electronegative atom bearing a lone pair of electrons—the hydrogen bond acceptor (Ac). Such an interacting system is generally denoted , where the solid line denotes a polar covalent bond, and the dotted or dashed line indicates the hydrogen bond. The most frequent donor and acceptor atoms are the second-row elements nitrogen (N), oxygen (O), and fluorine (F). Hydrogen bonds can be intermolecular (occurring between separate molecules) or intramolecular (occurring among parts of the same molecule). The energy of a hydrogen bond depends on the geometry, the environment, and the nature of the specific donor and acceptor atoms and can vary between 1 and 40 kcal/mol. This makes them somewhat stronger than a van der Waals interaction, and weaker than fully covalent ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Urea
Urea, also known as carbamide, is an organic compound with chemical formula . This amide has two amino groups (–) joined by a carbonyl functional group (–C(=O)–). It is thus the simplest amide of carbamic acid. Urea serves an important role in the metabolism of nitrogen-containing compounds by animals and is the main nitrogen-containing substance in the urine of mammals. It is a colorless, odorless solid, highly soluble in water, and practically non-toxic ( is 15 g/kg for rats). Dissolved in water, it is neither acidic nor alkaline. The body uses it in many processes, most notably nitrogen excretion. The liver forms it by combining two ammonia molecules () with a carbon dioxide () molecule in the urea cycle. Urea is widely used in fertilizers as a source of nitrogen (N) and is an important raw material for the chemical industry. In 1828 Friedrich Wöhler discovered that urea can be produced from inorganic starting materials, which was an important conceptual milestone ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Xylose Isomerase
In enzymology, a xylose isomerase () is an enzyme that catalyzes the interconversion of D-xylose and D-xylulose. This enzyme belongs to the family of isomerases, specifically those intramolecular oxidoreductases interconverting aldoses and ketoses. The isomerase has now been observed in nearly a hundred species of bacteria. Xylose-isomerases are also commonly called fructose-isomerases due to their ability to interconvert glucose and fructose. The systematic name of this enzyme class is D-xylose aldose-ketose-isomerase. Other names in common use include D-xylose isomerase, D-xylose ketoisomerase, and D-xylose ketol-isomerase. History The activity of D-xylose isomerase was first observed by Mitsuhashi and Lampen in 1953 in the bacterium ''Lactobacillus pentosus''. Artificial production through transformed ''E.coli'' have also been successful. In 1957, the D-xylose isomerase activity on D-glucose conversion to D-fructose was noted by Kooi and Marshall. It is now known that isome ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pseudomonas
''Pseudomonas'' is a genus of Gram-negative, Gammaproteobacteria, belonging to the family Pseudomonadaceae and containing 191 described species. The members of the genus demonstrate a great deal of metabolic diversity and consequently are able to colonize a wide range of niches. Their ease of culture ''in vitro'' and availability of an increasing number of ''Pseudomonas'' strain genome sequences has made the genus an excellent focus for scientific research; the best studied species include ''P. aeruginosa'' in its role as an opportunistic human pathogen, the plant pathogen '' P. syringae'', the soil bacterium '' P. putida'', and the plant growth-promoting ''P. fluorescens, P. lini, P. migulae'', and ''P. graminis''. Because of their widespread occurrence in water and plant seeds such as dicots, the pseudomonads were observed early in the history of microbiology. The generic name ''Pseudomonas'' created for these organisms was defined in rather vague terms by Walter Migula i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
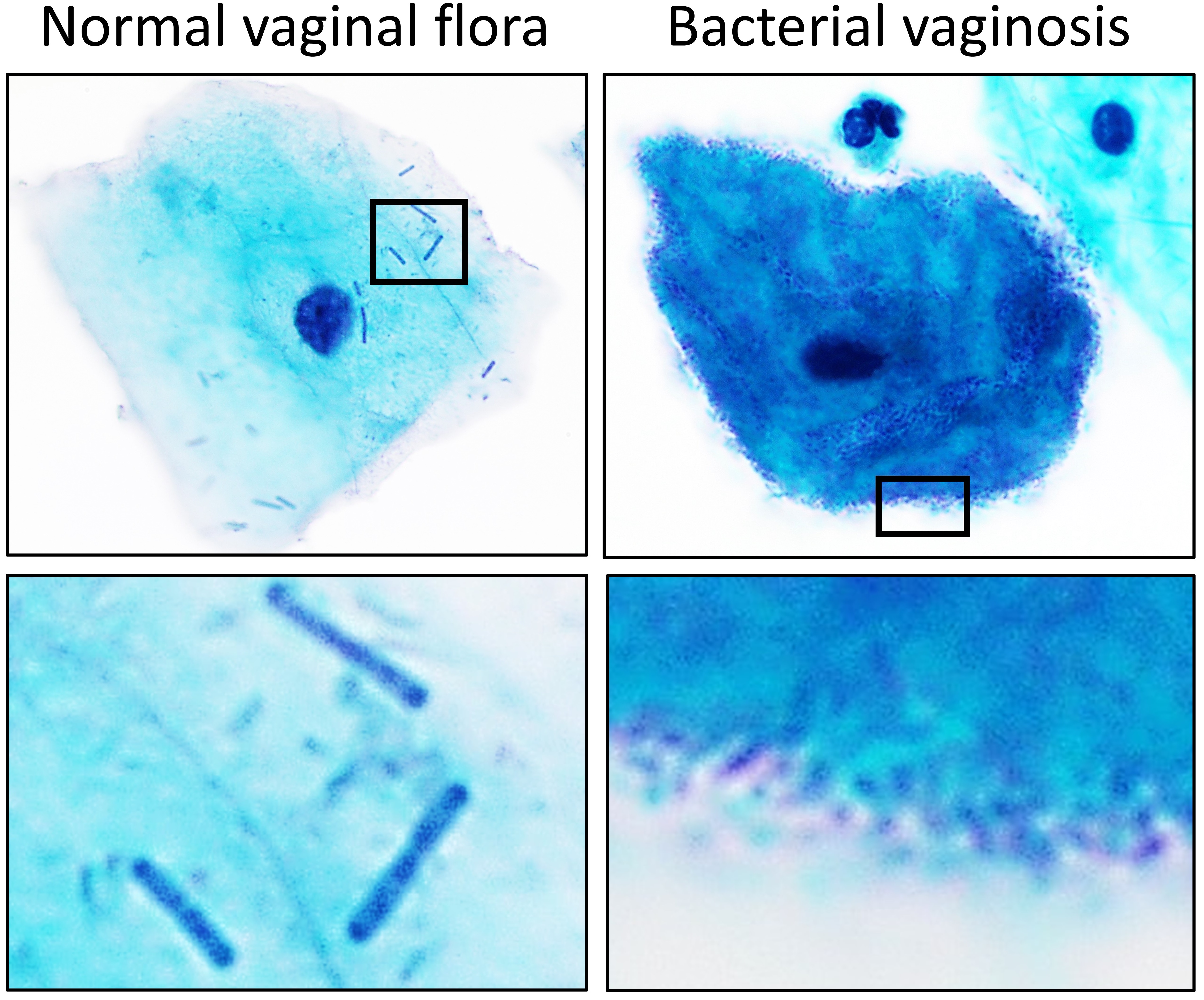
Lactobacillus
''Lactobacillus'' is a genus of Gram-positive, aerotolerant anaerobes or microaerophilic, rod-shaped, non-spore-forming bacteria. Until 2020, the genus ''Lactobacillus'' comprised over 260 phylogenetically, ecologically, and metabolically diverse species; a taxonomic revision of the genus assigned lactobacilli to 25 genera (see below). ''Lactobacillus'' species constitute a significant component of the human and animal microbiota at a number of body sites, such as the digestive system, and the female genital system. In women of European ancestry, ''Lactobacillus'' species are normally a major part of the vaginal microbiota. ''Lactobacillus'' forms biofilms in the vaginal and gut microbiota, allowing them to persist during harsh environmental conditions and maintain ample populations. ''Lactobacillus'' exhibits a mutualistic relationship with the human body, as it protects the host against potential invasions by pathogens, and in turn, the host provides a source of nutrien ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Azotobacter
''Azotobacter'' is a genus of usually motile, oval or spherical bacteria that form thick-walled cysts (and also has hard crust) and may produce large quantities of capsular slime. They are aerobic, free-living soil microbes that play an important role in the nitrogen cycle in nature, binding atmospheric nitrogen, which is inaccessible to plants, and releasing it in the form of ammonium ions into the soil (nitrogen fixation). In addition to being a model organism for studying diazotrophs, it is used by humans for the production of biofertilizers, food additives, and some biopolymers. The first representative of the genus, ''Azotobacter chroococcum'', was discovered and described in 1901 by Dutch microbiologist and botanist Martinus Beijerinck. ''Azotobacter'' species are Gram-negative bacteria found in neutral and alkaline soils, in water, and in association with some plants. Biological characteristics Morphology Cells of the genus ''Azotobacter'' are relatively large for b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |