|
H5N1 Genetic Structure
H5N1 genetic structure is the molecular structure of the H5N1 virus's RNA. H5N1 is an Influenza A virus subtype. Experts believe it might mutate into a form that transmits easily from person to person. If such a mutation occurs, it might remain an H5N1 subtype or could shift subtypes as did H2N2 when it evolved into the Hong Kong Flu strain of H3N2. H5N1 has mutatedFigure 1 of the article gives a diagramatic representation of the genetic relatedness of Asian H5N1 genes from various isolates of the virus. through into dozens of highly |
H5N1
Influenza A virus subtype H5N1 (A/H5N1) is a subtype of the influenza A virus which can cause illness in humans and many other animal species. A bird-adapted strain of H5N1, called HPAI A(H5N1) for highly pathogenic avian influenza virus of type A of subtype H5N1, is the highly pathogenic causative agent of H5N1 flu, commonly known as avian influenza ("bird flu"). It is enzootic (maintained in the population) in many bird populations, especially in Southeast Asia. One strain of HPAI A(H5N1) is spreading globally after first appearing in Asia. It is epizootic (an epidemic in nonhumans) and panzootic (affecting animals of many species, especially over a wide area), killing tens of millions of birds and spurring the culling of hundreds of millions of others to stem its spread. Many references to "bird flu" and H5N1 in the popular media refer to this strain. This was reprinted in 2005: According to the World Health Organization (WHO) and the United Nations Food and Agriculture O ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antigen
In immunology, an antigen (Ag) is a molecule or molecular structure or any foreign particulate matter or a pollen grain that can bind to a specific antibody or T-cell receptor. The presence of antigens in the body may trigger an immune response. The term ''antigen'' originally referred to a substance that is an antibody generator. Antigens can be proteins, peptides (amino acid chains), polysaccharides (chains of monosaccharides/simple sugars), lipids, or nucleic acids. Antigens are recognized by antigen receptors, including antibodies and T-cell receptors. Diverse antigen receptors are made by cells of the immune system so that each cell has a specificity for a single antigen. Upon exposure to an antigen, only the lymphocytes that recognize that antigen are activated and expanded, a process known as clonal selection. In most cases, an antibody can only react to and bind one specific antigen; in some instances, however, antibodies may cross-react and bind more than one antigen. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Virulence
Virulence is a pathogen's or microorganism's ability to cause damage to a host. In most, especially in animal systems, virulence refers to the degree of damage caused by a microbe to its host. The pathogenicity of an organism—its ability to cause disease—is determined by its virulence factors. In the specific context of gene for gene systems, often in plants, virulence refers to a pathogen's ability to infect a resistant host. The noun ''virulence'' derives from the adjective ''virulent'', meaning disease severity. The word ''virulent'' derives from the Latin word ''virulentus'', meaning "a poisoned wound" or "full of poison." From an ecological standpoint, virulence is the loss of fitness induced by a parasite upon its host. Virulence can be understood in terms of proximate causes—those specific traits of the pathogen that help make the host ill—and ultimate causes—the evolutionary pressures that lead to virulent traits occurring in a pathogen strain. Virulent ba ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template. Using the enzyme helicase, RNAP locally opens the double-stranded DNA so that one strand of the exposed nucleotides can be used as a template for the synthesis of RNA, a process called transcription. A transcription factor and its associated transcription mediator complex must be attached to a DNA binding site called a promoter region before RNAP can initiate the DNA unwinding at that position. RNAP not only initiates RNA transcription, it also guides the nucleotides into position, facilitates attachment and elongation, has intrinsic proofreading and replacement capabilities, and termination recognition capability. In eukaryotes, RNAP can build chains as long as 2.4 million nucleotides. RNAP produces RNA that, functionally, is either for protein coding, i.e. messenger RNA (mRNA); or n ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Basic Reproduction Number
In epidemiology, the basic reproduction number, or basic reproductive number (sometimes called basic reproduction ratio or basic reproductive rate), denoted R_0 (pronounced ''R nought'' or ''R zero''), of an infection is the expected number of cases directly generated by one case in a population where all individuals are susceptible to infection. The definition assumes that no other individuals are infected or immunized (naturally or through vaccination). Some definitions, such as that of the Australian Department of Health, add the absence of "any deliberate intervention in disease transmission". The basic reproduction number is not necessarily the same as the effective reproduction number R (usually written R_t 't'' for time sometimes R_e), which is the number of cases generated in the current state of a population, which does not have to be the uninfected state. R_0 is a dimensionless number (persons infected per person infecting) and not a time rate, which would have units ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
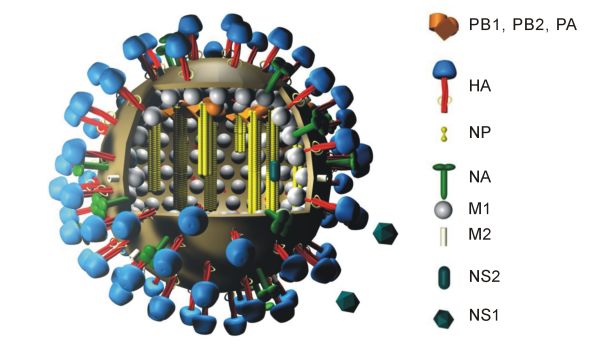
NS1 Influenza Protein
The NS1 influenza protein (NS1) is a viral nonstructural protein encoded by the NS gene segments of type Influenza A virus, A, Influenza B, B and Influenza C virus, C influenza viruses. Also encoded by this segment is the nuclear export protein (NEP), formally referred to as NS2 (HCV), NS2 protein, which mediates the export of influenza virus ribonucleoprotein (RNP) complexes from the nucleus, where they are assembled.Influenza B and C Virus NEP (NS2) Proteins Possess Nuclear Export Activities Journal of Virology, August 2001, p. 7375-7383, Vol. 75, No. 16 Characteristics The NS1 of influenza A virus is a 26,000 Dalton protein. It prevents polyadenylation of cellular mRNAs to circumvent antiviral responses of the host, e ...[...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
M2 Protein
The Matrix-2 (M2) protein is a proton-selective viroporin, integral in the viral envelope of the influenza A virus. The channel itself is a homotetramer (consists of four identical M2 units), where the units are helices stabilized by two disulfide bonds, and is activated by low pH. The M2 protein is encoded on the seventh RNA segment together with the M1 protein. Proton conductance by the M2 protein in influenza A is essential for viral replication. Influenza B and C viruses encode proteins with similar function dubbed "BM2" and "CM2" respectively. They share little similarity with M2 at the sequence level, despite a similar overall structure and mechanism. Structure In influenza A virus, M2 protein unit consists of three protein segments comprising 97 amino acid residues: (i) an extracellular N-terminal domain (residues 1–23); (ii) a transmembrane segment (TMS) (residues 24–46); (iii) an intracellular C-terminal domain (residues 47–97). The TMS forms the pore of the ion ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
M1 Protein
The M1 protein is a matrix protein of the influenza virus. It forms a coat inside the viral envelope. This is a bifunctional membrane/RNA-binding protein that mediates the encapsidation of nucleoprotein cores into the membrane envelope. It is therefore required that M1 binds both membrane and RNA simultaneously. The M1 protein binds to the viral RNA. The binding is not specific to any RNA sequence, and is performed via a peptide sequence rich in basic amino acids. It also has multiple regulatory functions, performed by interaction with the components of the host cell. The mechanisms regulated include a role in the export of the viral ribonucleoproteins from the host cell nucleus, inhibition of viral transcription, and a role in the virus assembly and budding. The protein was found to undergo phosphorylation in the host cell. The M1 protein forms a layer under the patches of host cell membrane that are rich with the viral hemagglutinin, neuraminidase and M2 transmembrane proteins ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Viral Neuraminidase
Viral neuraminidase is a type of neuraminidase found on the surface of influenza viruses that enables the virus to be released from the host cell. Neuraminidases are enzymes that cleave sialic acid (also called neuraminic acid) groups from glycoproteins. Neuraminidase inhibitors are antiviral agents that inhibit influenza viral neuraminidase activity and are of major importance in the control of influenza. Viral neuraminidases are the members of the glycoside hydrolase family 3CAZY GH_34which comprises enzymes with only one known activity; sialidase or neuraminidase . Neuraminidases cleave the terminal sialic acid residues from carbohydrate chains in glycoproteins. Sialic acid is a negatively charged sugar associated with the protein and lipid portions of lipoproteins. To infect a host cell, the influenza virus attaches to the exterior cell surface using hemagglutinin, a molecule found on the surface of the virus that binds to sialic acid groups. Sialic acids are found on v ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Influenza Virus Nucleoprotein
Influenza virus nucleoprotein (NP) is a structural protein which encapsidates the negative strand viral RNA. NP is one of the main determinants of species specificity. The question of how far the NP gene can cross the species barrier by reassortment and become adapted by mutation In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mi ... to the new host has been discussed. References {{DEFAULTSORT:Influenza Virus Nucleoprotein Structural proteins Influenza ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hemagglutinin (influenza)
Influenza hemagglutinin (HA) or haemagglutinin .html" ;"title="/sup>">/sup> (British English) is a homotrimeric glycoprotein found on the surface of influenza viruses and is integral to its infectivity. Hemagglutinin is a Class I Fusion Protein, having multifunctional activity as both an attachment factor and membrane fusion protein. Therefore, HA is responsible for binding Influenza virus to sialic acid on the surface of target cells, such as cells in the upper respiratory tract or erythrocytes, causing as a result the internalization of the virus. Secondarily, HA is responsible for the fusion of the viral envelope with the late endosomal membrane once exposed to low pH (5.0-5.5). The name "hemagglutinin" comes from the protein's ability to cause red blood cells (erythrocytes) to clump together (" agglutinate") ''in vitro''. Subtypes Hemagglutinin (HA) in influenza A has at least 18 different subtypes. These subtypes are named H1 through H18. H16 was discovered in 2004 on in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Open Reading Frame
In molecular biology, open reading frames (ORFs) are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a prokaryotic DNA sequence, where only one of the six possible reading frames will be "open" (the "reading", however, refers to the RNA produced by transcription of the DNA and its subsequent interaction with the ribosome in translation). Such an ORF may contain a start codon (usually AUG in terms of RNA) and by definition cannot extend beyond a stop codon (usually UAA, UAG or UGA in RNA). That start codon (not necessarily the first) indicates where translation may start. The transcription termination site is located after the ORF, beyond the translation stop codon. If transcription were to cease before the stop codon, an incomplete protein would be made during translation. In eukaryotic genes with multiple exons, introns are removed and exons are then joined together after transcription to yield the final ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |