|
M1 Protein
The M1 protein is a matrix protein of the influenza virus. It forms a coat inside the viral envelope. This is a bifunctional membrane/RNA-binding protein that mediates the encapsidation of nucleoprotein cores into the membrane envelope. It is therefore required that M1 binds both membrane and RNA simultaneously. The M1 protein binds to the viral RNA. The binding is not specific to any RNA sequence, and is performed via a peptide sequence rich in basic amino acids. It also has multiple regulatory functions, performed by interaction with the components of the host cell. The mechanisms regulated include a role in the export of the viral ribonucleoproteins from the host cell nucleus, inhibition of viral transcription, and a role in the virus assembly and budding. The protein was found to undergo phosphorylation in the host cell. The M1 protein forms a layer under the patches of host cell membrane that are rich with the viral hemagglutinin, neuraminidase and M2 transmembrane proteins ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Matrix Protein
Viral matrix proteins are structural proteins linking the viral envelope with the virus core. They play a crucial role in virus assembly, and interact with the RNP complex as well as with the viral membrane. They are found in many enveloped viruses including paramyxoviruses, orthomyxoviruses, herpesviruses, retroviruses, filoviruses and other groups. An example is the M1 protein of the influenza virus, showing affinity to the glycoproteins inserted in the host cell membrane on one side and affinity for the RNP complex molecules on the other side, which allows formation at the membrane of a complex made of the viral ribonucleoprotein at the inner side indirectly connected to the viral glycoproteins protruding from the membrane. This assembly complex will now bud out of the cell as new mature viruses. Viral matrix proteins, like many other viral proteins, can exert different functions during the course of the infection. For example, in rhabdoviruses, binding of M proteins to nucle ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Budding
Budding or blastogenesis is a type of asexual reproduction in which a new organism develops from an outgrowth or bud due to cell division at one particular site. For example, the small bulb-like projection coming out from the yeast cell is known as a bud. Since the reproduction is asexual, the newly created organism is a clone and excepting mutations is genetically identical to the parent organism. Organisms such as hydra use regenerative cells for reproduction in the process of budding. In hydra, a bud develops as an outgrowth due to repeated cell division at one specific site. These buds develop into tiny individuals and, when fully mature, detach from the parent body and become new independent individuals. Internal budding or endodyogeny is a process of asexual reproduction, favored by parasites such as ''Toxoplasma gondii''. It involves an unusual process in which two daughter cells are produced inside a mother cell, which is then consumed by the offspring prior to their s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Membrane Biology
Membrane biology is the study of the biological and physiochemical characteristics of membranes, with applications in the study of cellular physiology. Membrane bioelectrical impulses are described by the Hodgkin cycle. Biophysics Membrane biophysics is the study of biological membrane structure and function using physical, computational, mathematical, and biophysical methods. A combination of these methods can be used to create phase diagrams of different types of membranes, which yields information on thermodynamic behavior of a membrane and its components. As opposed to membrane biology, membrane biophysics focuses on quantitative information and modeling of various membrane phenomena, such as lipid raft The plasma membranes of cells contain combinations of glycosphingolipids, cholesterol and protein receptors organised in glycolipoprotein lipid microdomains termed lipid rafts. Their existence in cellular membranes remains somewhat controversial. ... formation, rates of l ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
H5N1 Genetic Structure
H5N1 genetic structure is the molecular structure of the H5N1 virus's RNA. H5N1 is an Influenza A virus subtype. Experts believe it might mutate into a form that transmits easily from person to person. If such a mutation occurs, it might remain an H5N1 subtype or could shift subtypes as did H2N2 when it evolved into the Hong Kong Flu strain of H3N2. H5N1 has mutatedFigure 1 of the article gives a diagramatic representation of the genetic relatedness of Asian H5N1 genes from various isolates of the virus. through into dozens of highly |
Cis-regulatory Element
''Cis''-regulatory elements (CREs) or ''Cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology. CREs are found in the vicinity of the genes that they regulate. CREs typically regulate gene transcription by binding to transcription factors. A single transcription factor may bind to many CREs, and hence control the expression of many genes ( pleiotropy). The Latin prefix ''cis'' means "on this side", i.e. on the same molecule of DNA as the gene(s) to be transcribed. CRMs are stretches of DNA, usually 100–1000 DNA base pairs in length, where a number of transcription factors can bind and regulate expression of nearby genes and regulate their transcription rates. They are labeled as ''cis'' because they are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helix
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence. The alpha helix is also called a classic Pauling–Corey–Branson α-helix. The name 3.613-helix is also used for this type of helix, denoting the average number of residues per helical turn, with 13 atoms being involved in the ring formed by the hydrogen bond. Among types of local structure in proteins, the α-helix is the most extreme and the most predictable from sequence, as well as the most prevalent. Discovery In the early 1930s, William Astbury showed that there were drastic changes in the X-ray fiber diffraction of moist wool or hair fibers upon significant stretching. The data suggested that the unstretched fibers had a coiled molecular structure with a characteristic repeat of ≈. Astb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the three dimensional conformational isomerism, form of ''local segments'' of proteins. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Domain (biology)
In biological taxonomy, a domain ( or ) (Latin: ''regio''), also dominion, superkingdom, realm, or empire, is the highest taxonomic rank of all organisms taken together. It was introduced in the three-domain system of taxonomy devised by Carl Woese, Otto Kandler and Mark Wheelis in 1990. According to the domain system, the tree of life consists of either three domains such as Archaea, Bacteria, and Eukarya, or two domains consisting of Archaea and Bacteria, with Eukarya included in Archaea. The first two are all prokaryotes, single-celled microorganisms without a membrane-bound nucleus. All organisms that have a cell nucleus and other membrane-bound organelles are included in Eukarya. Non-cellular life is not included in this system. Alternatives to the three-domain system include the earlier two-empire system (with the empires Prokaryota and Eukaryota), and the eocyte hypothesis (with two domains of Bacteria and Archaea, with Eukarya included as a branch of Archaea). Term ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence (biology)
A sequence in biology is the one-dimensional ordering of monomers, covalently linked within a biopolymer; it is also referred to as the primary structure of a biological macromolecule. While it can refer to many different molecules, the term sequence is most often used to refer to a DNA sequence. See also * Protein sequence * DNA sequence * Genotype * Self-incompatibility in plants * List of geneticists * Human Genome Project * Dot plot (bioinformatics) * Multiplex Ligation-dependent Probe Amplification * Sequence analysis In bioinformatics, sequence analysis is the process of subjecting a DNA, RNA or peptide sequence to any of a wide range of analytical methods to understand its features, function, structure, or evolution. Methodologies used include sequence alig ... Molecular biology {{molecular-biology-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Domain
In molecular biology, a protein domain is a region of a protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins. Background The concept of the domain was first proposed in 1973 by Wetlaufer aft ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
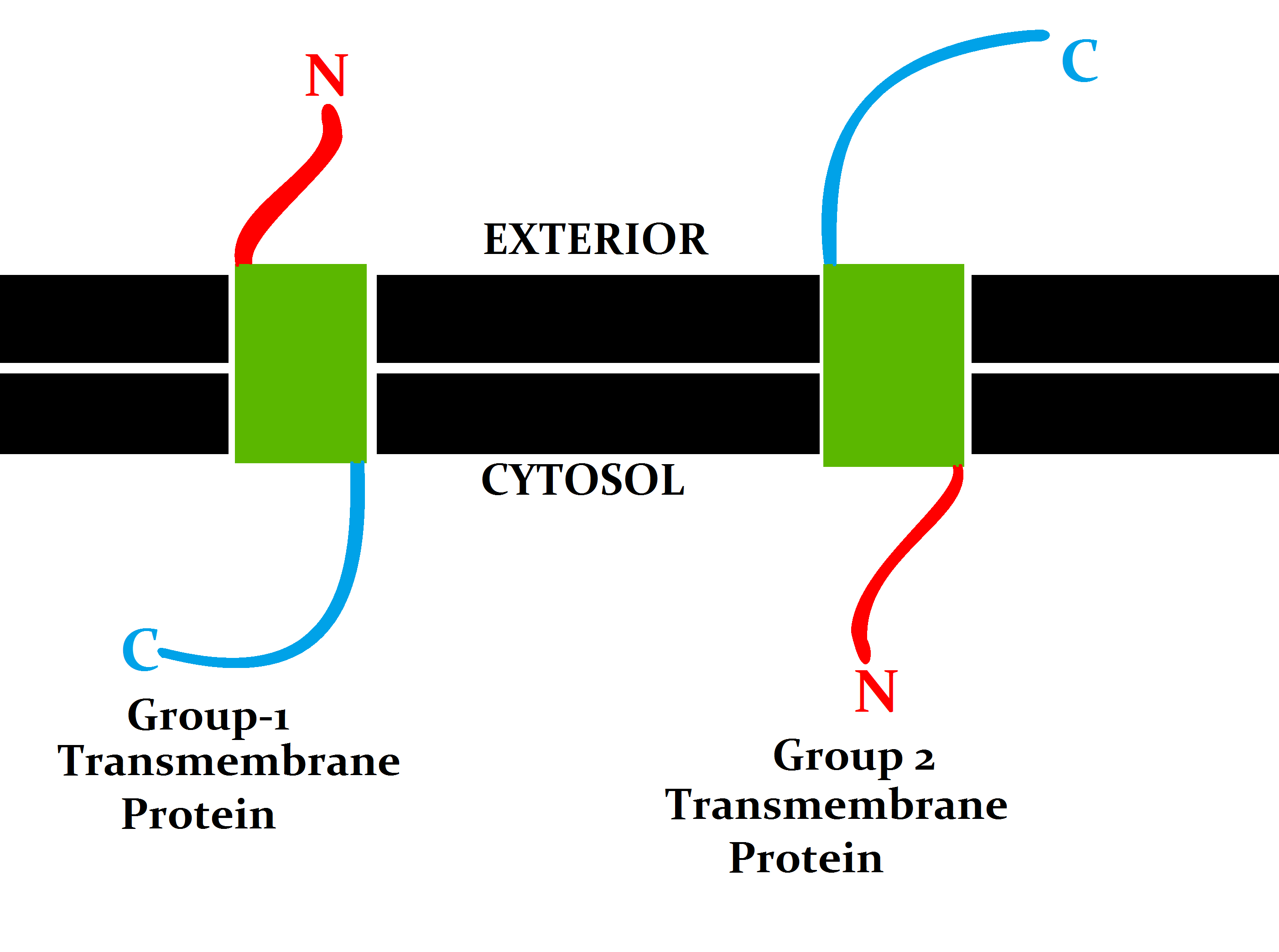
Transmembrane Protein
A transmembrane protein (TP) is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents. The peptide sequence that spans the membrane, or the transmembrane segment, is largely hydrophobic and can be visualized using the hydropathy plot. Depending on the number of transmembrane segments, transmembrane proteins can be classified as single-span (or bitopic) or multi-span (polytopic). Some other integral membrane proteins are called monotopic, meaning that they are also permanently attached to the membrane, but do not pass ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
M2 Protein
The Matrix-2 (M2) protein is a proton-selective viroporin, integral in the viral envelope of the influenza A virus. The channel itself is a homotetramer (consists of four identical M2 units), where the units are helices stabilized by two disulfide bonds, and is activated by low pH. The M2 protein is encoded on the seventh RNA segment together with the M1 protein. Proton conductance by the M2 protein in influenza A is essential for viral replication. Influenza B and C viruses encode proteins with similar function dubbed "BM2" and "CM2" respectively. They share little similarity with M2 at the sequence level, despite a similar overall structure and mechanism. Structure In influenza A virus, M2 protein unit consists of three protein segments comprising 97 amino acid residues: (i) an extracellular N-terminal domain (residues 1–23); (ii) a transmembrane segment (TMS) (residues 24–46); (iii) an intracellular C-terminal domain (residues 47–97). The TMS forms the pore of the ion ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |