|
HTERT
Telomerase reverse transcriptase (abbreviated to TERT, or hTERT in humans) is a catalytic subunit of the enzyme telomerase, which, together with the telomerase RNA component (TERC), comprises the most important unit of the telomerase complex. Telomerases are part of a distinct subgroup of RNA-dependent polymerases. Telomerase lengthens telomeres in DNA strands, thereby allowing senescent cells that would otherwise become postmitotic and undergo apoptosis to exceed the Hayflick limit and become potentially immortal, as is often the case with cancerous cells. To be specific, TERT is responsible for catalyzing the addition of nucleotides in a TTAGGG sequence to the ends of a chromosome's telomeres. This addition of repetitive DNA sequences prevents degradation of the chromosomal ends following multiple rounds of replication. hTERT absence (usually as a result of a chromosomal mutation) is associated with the disorder Cri du chat. Function Telomerase is a ribonucleoprotein p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Telomerase
Telomerase, also called terminal transferase, is a ribonucleoprotein that adds a species-dependent telomere repeat sequence to the 3' end of telomeres. A telomere is a region of repetitive sequences at each end of the chromosomes of most eukaryotes. Telomeres protect the end of the chromosome from DNA damage or from fusion with neighbouring chromosomes. The fruit fly ''Drosophila melanogaster'' lacks telomerase, but instead uses retrotransposons to maintain telomeres. Telomerase is a reverse transcriptase enzyme that carries its own RNA molecule (e.g., with the sequence 3′- CCC AA UCCC-5′ in ''Trypanosoma brucei'') which is used as a template when it elongates telomeres. Telomerase is active in gametes and most cancer cells, but is normally absent from, or at very low levels in, most somatic cells. History The existence of a compensatory mechanism for telomere shortening was first found by Soviet biologist Alexey Olovnikov in 1973, who also suggested the telomere hypothe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Telomerase
Telomerase, also called terminal transferase, is a ribonucleoprotein that adds a species-dependent telomere repeat sequence to the 3' end of telomeres. A telomere is a region of repetitive sequences at each end of the chromosomes of most eukaryotes. Telomeres protect the end of the chromosome from DNA damage or from fusion with neighbouring chromosomes. The fruit fly ''Drosophila melanogaster'' lacks telomerase, but instead uses retrotransposons to maintain telomeres. Telomerase is a reverse transcriptase enzyme that carries its own RNA molecule (e.g., with the sequence 3′- CCC AA UCCC-5′ in ''Trypanosoma brucei'') which is used as a template when it elongates telomeres. Telomerase is active in gametes and most cancer cells, but is normally absent from, or at very low levels in, most somatic cells. History The existence of a compensatory mechanism for telomere shortening was first found by Soviet biologist Alexey Olovnikov in 1973, who also suggested the telomere hypothe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cri Du Chat
Cri du chat syndrome is a rare genetic disorder due to a partial chromosome deletion on chromosome 5. Its name is a French term ("cat-cry" or " call of the cat") referring to the characteristic cat-like cry of affected children. It was first described by Jérôme Lejeune in 1963. The condition affects an estimated 1 in 50,000 live births across all ethnicities and is more common in females by a 4:3 ratio. Signs and symptoms The syndrome gets its name from the characteristic cry of affected infants, which is similar to that of a meowing kitten, due to problems with the larynx and nervous system. About one third of children lose the cry by age of 2 years. Other symptoms of cri du chat syndrome may include: * feeding problems because of difficulty in swallowing and sucking; * mutism; * low birth weight and poor growth; * severe cognitive, speech and motor disabilities; * behavioural problems such as hyperactivity, aggression, outbursts and repetitive movements; * unusual facial fe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Telomerase RNA Component
Telomerase RNA component, also known as TR, TER or TERC, is an ncRNA found in eukaryotes that is a component of telomerase, the enzyme used to extend telomeres. TERC serves as a template for telomere replication ( reverse transcription) by telomerase. Telomerase RNAs differ greatly in sequence and structure between vertebrates, ciliates and yeasts, but they share a 5' pseudoknot structure close to the template sequence. The vertebrate telomerase RNAs have a 3' H/ACA snoRNA-like domain. Structure TERC is a Long non-coding RNA (lncRNA) ranging in length from ~150nt in ciliates to 400-600nt in vertebrates, and 1,300nt in yeast (Alnafakh). Mature human TERC (hTR) is 451nt in length. TERC has extensive secondary structural features over 4 principal conserved domains. The core domain, the largest domain at the 5’ end of TERC, contains the CUAAC Telomere template sequence. Its secondary structure consists of a large loop containing the template sequence, a P1 loop-closing heli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
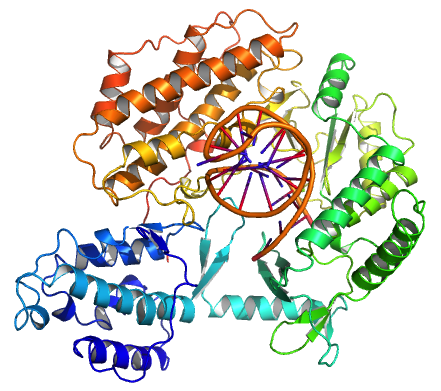
Sp1 Transcription Factor
Transcription factor Sp1, also known as specificity protein 1* is a protein that in humans is encoded by the SP1 gene. Function The protein encoded by this gene is a zinc finger transcription factor that binds to GC-rich motifs of many promoters. The encoded protein is involved in many cellular processes, including cell differentiation, cell growth, apoptosis, immune responses, response to DNA damage, and chromatin remodeling. Post-translational modifications such as phosphorylation, acetylation, ''O''-GlcNAcylation, and proteolytic processing significantly affect the activity of this protein, which can be an activator or a repressor. In the SV40 virus, Sp1 binds to the GC boxes in the regulatory region (RR) of the genome. Structure SP1 belongs to the Sp/KLF family of transcription factors. The protein is 785 amino acids long, with a molecular weight of 81 kDa. The SP1 transcription factor contains two glutamine-rich activation domains at its N-terminus that are believ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-Myc
''Myc'' is a family of regulator genes and proto-oncogenes that code for transcription factors. The ''Myc'' family consists of three related human genes: ''c-myc'' (MYC), ''l-myc'' (MYCL), and ''n-myc'' (MYCN). ''c-myc'' (also sometimes referred to as ''MYC'') was the first gene to be discovered in this family, due to homology with the viral gene ''v-myc''. In cancer, ''c-myc'' is often constitutively (persistently) expressed. This leads to the increased expression of many genes, some of which are involved in cell proliferation, contributing to the formation of cancer. A common human translocation involving ''c-myc'' is critical to the development of most cases of Burkitt lymphoma. Constitutive upregulation of ''Myc'' genes have also been observed in carcinoma of the cervix, colon, breast, lung and stomach. Myc is thus viewed as a promising target for anti-cancer drugs. Unfortunately, Myc possesses several features that render it undruggable such that any anti-cancer drugs f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Exons
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequence in RNA transcripts. In RNA splicing, introns are removed and exons are covalently joined to one another as part of generating the mature RNA. Just as the entire set of genes for a species constitutes the genome, the entire set of exons constitutes the exome. History The term ''exon'' derives from the expressed region and was coined by American biochemist Walter Gilbert in 1978: "The notion of the cistron… must be replaced by that of a transcription unit containing regions which will be lost from the mature messengerwhich I suggest we call introns (for intragenic regions)alternating with regions which will be expressedexons." This definition was originally made for protein-coding transcripts that are spliced before being translated. Th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Introns
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e. a region inside a gene."The notion of the cistron .e., gene... must be replaced by that of a transcription unit containing regions which will be lost from the mature messenger – which I suggest we call introns (for intragenic regions) – alternating with regions which will be expressed – exons." (Gilbert 1978) The term ''intron'' refers to both the DNA sequence within a gene and the corresponding RNA sequence in RNA transcripts. The non-intron sequences that become joined by this RNA processing to form the mature RNA are called exons. Introns are found in the genes of most organisms and many viruses and they can be located in both protein-coding genes and genes that function as RNA (noncoding genes). There are four main types of introns: tRNA introns, group I introns, group II introns, and s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TATA Box
In molecular biology, the TATA box (also called the Goldberg–Hogness box) is a sequence of DNA found in the core promoter region of genes in archaea and eukaryotes. The bacterial homolog of the TATA box is called the Pribnow box which has a shorter consensus sequence. The TATA box is considered a non-coding DNA sequence (also known as a cis-regulatory element). It was termed the "TATA box" as it contains a consensus sequence characterized by repeating T and A base pairs. How the term "box" originated is unclear. In the 1980s, while investigating nucleotide sequences in mouse genome loci, the Hogness box sequence was found and "boxed in" at the -31 position. When consensus nucleotides and alternative ones were compared, homologous regions were "boxed" by the researchers. The boxing in of sequences sheds light on the origin of the term "box". The TATA box was first identified in 1978 as a component of eukaryotic promoters. Transcription is initiated at the TATA box in TAT ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factors
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization (body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment of RNA po ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |