|
GFF3
In bioinformatics, the general feature format (gene-finding format, generic feature format, GFF) is a file format used for describing genes and other features of DNA, RNA and protein sequences. GFF Versions The following versions of GFF exist: General Feature Format Version 2 generally deprecated * a derivative used by Ensembl Generic Feature Format Version 3*Genome Variation Format with additional pragmas and attributes for sequence_alteration features GFF2/GTF had a number of deficiencies, notably that it can only represent two-level feature hierarchies and thus cannot handle the three-level hierarchy of gene → transcript → exon. GFF3 addresses this and other deficiencies. For example, it supports arbitrarily many hierarchical levels, and gives specific meanings to certain tags in the attributes field. The GTF is identical to GFF, version 2. GFF general structure All GFF formats (GFF2, GFF3 and GTF) are tab delimited with 9 fields per line. They all share the same stru ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
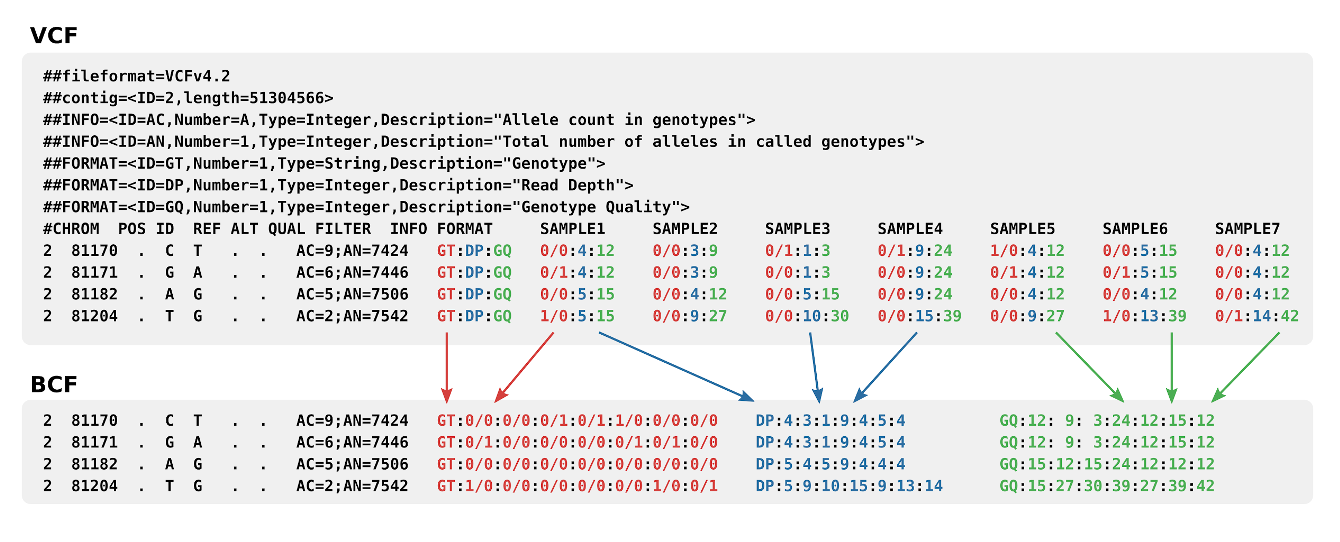
Variant Call Format
The Variant Call Format (VCF) specifies the format of a text file used in bioinformatics for storing gene sequence variations. The format has been developed with the advent of large-scale genotyping and DNA sequencing projects, such as the 1000 Genomes Project. Existing formats for genetic data such as General feature format, General feature format (GFF) stored all of the genetic data, much of which is redundant because it will be shared across the genomes. By using the variant call format only the variations need to be stored along with a reference genome. The standard is currently in version 4.3, although the 1000 Genomes Project has developed its own specification for structural variations such as duplications, which are not easily accommodated into the existing schema. There is also a genomic VCF (gVCF) extended format, which includes additional information about "blocks" that match the reference and their qualities. A set of tools is also available for editing and manipulati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bioinformatics
Bioinformatics () is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combines biology, chemistry, physics, computer science, information engineering, mathematics and statistics to analyze and interpret the biological data. Bioinformatics has been used for '' in silico'' analyses of biological queries using computational and statistical techniques. Bioinformatics includes biological studies that use computer programming as part of their methodology, as well as specific analysis "pipelines" that are repeatedly used, particularly in the field of genomics. Common uses of bioinformatics include the identification of candidates genes and single nucleotide polymorphisms (SNPs). Often, such identification is made with the aim to better understand the genetic basis of disease, unique adaptations, desirable properties (e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BED (file Format)
The BED (Browser Extensible Data) format is a text file format used to store genomic regions as coordinates and associated annotations. The data are presented in the form of columns separated by spaces or tabs. This format was developed during the Human Genome Project and then adopted by other sequencing projects. As a result of this increasingly wide use, this format had already become a ''de facto'' standard in bioinformatics before a formal specification was written. One of the advantages of this format is the manipulation of coordinates instead of nucleotide sequences, which optimizes the power and computation time when comparing all or part of genomes. In addition, its simplicity makes it easy to manipulate and read (or parsing) coordinates or annotations using word processing and scripting languages such as Python, Ruby or Perl or more specialized tools such as BEDTools. History The end of the 20th century saw the emergence of the first projects to sequence complete ge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are typically represented as rows within a matrix. Gaps are inserted between the residues so that identical or similar characters are aligned in successive columns. Sequence alignments are also used for non-biological sequences, such as calculating the distance cost between strings in a natural language or in financial data. Interpretation If two sequences in an alignment share a common ancestor, mismatches can be interpreted as point mutations and gaps as indels (that is, insertion or deletion mutations) introduced in one or both lineages in the time since they diverged from one another. In sequence alignments of proteins, the degree of similarity between amino acids occupying a parti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Distributed Annotation System
Distribution may refer to: Mathematics *Distribution (mathematics), generalized functions used to formulate solutions of partial differential equations * Probability distribution, the probability of a particular value or value range of a variable ** Cumulative distribution function, in which the probability of being no greater than a particular value is a function of that value *Frequency distribution, a list of the values recorded in a sample * Inner distribution, and outer distribution, in coding theory *Distribution (differential geometry), a subset of the tangent bundle of a manifold * Distributed parameter system, systems that have an infinite-dimensional state-space *Distribution of terms, a situation in which all members of a category are accounted for *Distributivity, a property of binary operations that generalises the distributive law from elementary algebra * Distribution (number theory) *Distribution problems, a common type of problems in combinatorics where the go ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ModENCODE
The Encyclopedia of DNA Elements (ENCODE) is a public research project which aims to identify functional elements in the human genome. ENCODE also supports further biomedical research by "generating community resources of genomics data, software, tools and methods for genomics data analysis, and products resulting from data analyses and interpretations." The current phase of ENCODE (2016-2019) is adding depth to its resources by growing the number of cell types, data types, assays and now includes support for examination of the mouse genome. History ENCODE was launched by the US National Human Genome Research Institute (NHGRI) in September 2003. Intended as a follow-up to the Human Genome Project, the ENCODE project aims to identify all functional elements in the human genome. The project involves a worldwide consortium of research groups, and data generated from this project can be accessed through public databases. The initial release of ENCODE was in 2013 and since has be ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Integrated Genome Browser
Integrated Genome Browser (IGB) (pronounced Ig-Bee) is an open-source genome browser, a visualization tool used to observe biologically-interesting patterns in genomic data sets, including sequence data, gene models, alignments, and data from DNA microarrays. History Integrated Genome Browser was first developed at Affymetrix for their scientists and public sector collaborators to visualize data from genome-wide tiling arrays. The first iterations of IGB were developed using funding from NIH awarded to company scientists Gregg Helt and Tom Gingeras. In 2004, Affymetrix released IGB as open source software, along with the Genoviz SDK, a graphics library for building genome browser applications. The first release of the code base was done as a compressed file archive. Soon after, the code was imported into a new repository at SourceForge. Since then, all development has proceeded in public under an open source model. In early 2008, a group led by former Affymetrix employee Ann Lo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
UniProt
UniProt is a freely accessible database of protein sequence and functional information, many entries being derived from genome sequencing projects. It contains a large amount of information about the biological function of proteins derived from the research literature. It is maintained by the UniProt consortium, which consists of several European bioinformatics organisations and a foundation from Washington, DC, United States. The UniProt consortium The UniProt consortium comprises the European Bioinformatics Institute (EBI), the Swiss Institute of Bioinformatics (SIB), and the Protein Information Resource (PIR). EBI, located at the Wellcome Trust Genome Campus in Hinxton, UK, hosts a large resource of bioinformatics databases and services. SIB, located in Geneva, Switzerland, maintains the ExPASy (Expert Protein Analysis System) servers that are a central resource for proteomics tools and databases. PIR, hosted by the National Biomedical Research Foundation (NBRF) at the Geor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GitHub
GitHub, Inc. () is an Internet hosting service for software development and version control using Git. It provides the distributed version control of Git plus access control, bug tracking, software feature requests, task management, continuous integration, and wikis for every project. Headquartered in California, it has been a subsidiary of Microsoft since 2018. It is commonly used to host open source software development projects. As of June 2022, GitHub reported having over 83 million developers and more than 200 million repositories, including at least 28 million public repositories. It is the largest source code host . History GitHub.com Development of the GitHub.com platform began on October 19, 2007. The site was launched in April 2008 by Tom Preston-Werner, Chris Wanstrath, P. J. Hyett and Scott Chacon after it had been made available for a few months prior as a beta release. GitHub has an annual keynote called GitHub Universe. Organizational ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sense (molecular Biology)
In molecular biology and genetics, the sense of a nucleic acid molecule, particularly of a strand of DNA or RNA, refers to the nature of the roles of the strand and its complement in specifying a sequence of amino acids. Depending on the context, sense may have slightly different meanings. For example, negative-sense strand of DNA is equivalent to the template strand, whereas the positive-sense strand is the non-template strand whose nucleotide sequence is equivalent to the sequence of the mRNA transcript. DNA sense Because of the complementary nature of base-pairing between nucleic acid polymers, a double-stranded DNA molecule will be composed of two strands with sequences that are reverse complements of each other. To help molecular biologists specifically identify each strand individually, the two strands are usually differentiated as the "sense" strand and the "antisense" strand. An individual strand of DNA is referred to as positive-sense (also positive (+) or simply sense) ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
The Arabidopsis Information Resource
The Arabidopsis Information Resource (TAIR) is a community resource and online model organism database of genetic and molecular biology data for the model plant ''Arabidopsis thaliana ''Arabidopsis thaliana'', the thale cress, mouse-ear cress or arabidopsis, is a small flowering plant native to Eurasia and Africa. ''A. thaliana'' is considered a weed; it is found along the shoulders of roads and in disturbed land. A winter a ...'', commonly known as mouse-ear cress. TAIR integrates information about the Arabidopsis genome, genes, gene products, natural variants, mutant alleles and plant phenotypes and research literature. Data in TAIR can be retrieved using simple and advanced searches, bulk query and download tools, and in collections of prepared text files. The Arabidopsis genome and annotations can be visualized using the interactive SeqViewer and GBrowse tools. TAIR’s biocurators are responsible for acquiring and integrating data from the research literature (functional ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |