|
F420H2DH Family
The H+-translocating F420H2 Dehydrogenase (F420H2DH) Family''(TC# 3.D.9)is a member of the Na+ transporting Mrp superfamily. A single F420H2 dehydrogenase (also referred to as F420H2:quinol oxidoreductase) from the methanogenic archaeon, ''Methanosarcina mazei'' Gö1, has been shown to be a redox driven proton pump. The F420H2DH of ''M. mazei'' has a molecular size of about 120 kDa and contains Fe-S clusters and FAD. A similar five-subunit enzyme has been isolated from '' Methanolobus tindarius''. The sulfate-reducing ''Archaeoglobus fulgidus'' (and several other archaea) also have this enzyme. Function Reduction of 2-hydroxyphenazine by F420H2DH is accompanied by the translocation of 1 H+ per 2 electrons transferred. Transport Reaction The overall vectorial reaction catalyzed by F420H2DH is : Reduced donor (2e−) + H+ (in) ⇌ oxidized acceptor (2e−) + H+ (out) Role in Methanogenesis ''Methanomassiliicoccus luminyensis'' has been isolated from the human gut and requ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mrp Superfamily
The Na+ Transporting Mrp Superfamily is a superfamily of integral membrane transport proteins. It includes the TC families:2.A.63- The Monovalent Cation (K+ or Na+):Proton Antiporter-3 (CPA3) Family3.D.1- The H+ or Na+-translocating NADH Dehyrogenase (NDH) Family3.D.9- The H+-translocating F420H2 Dehydrogenase (F420H2DH) Family Mrp of ''Bacillus subtilis'' is a 7 subunit Na+/H+ antiporter An antiporter (also called exchanger or counter-transporter) is a cotransporter and integral membrane protein involved in secondary active transport of two or more different molecules or ions across a phospholipid membrane such as the plasma memb ... comple(TC# 2.A.63.1.4) All subunits are homologous to the subunits in other members of this monovalent cation (K+ or Na+):proton antiporter-3 (CPA3) family as well as subunits in the archaeal hydrogenasesTC#s 3.D.1.4.1an3.D.1.4.2, which share several subunits with NADH dehydrogenase subunits (3.D.1). The largest subunits of the Mrp complex (MrpA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ATP Synthase
ATP synthase is a protein that catalyzes the formation of the energy storage molecule adenosine triphosphate (ATP) using adenosine diphosphate (ADP) and inorganic phosphate (Pi). It is classified under ligases as it changes ADP by the formation of P-O bond (phosphodiester bond). ATP synthase is a molecular machine. The overall reaction catalyzed by ATP synthase is: * ADP + Pi + 2H+out ATP + H2O + 2H+in The formation of ATP from ADP and Pi is energetically unfavorable and would normally proceed in the reverse direction. In order to drive this reaction forward, ATP synthase couples ATP synthesis during cellular respiration to an electrochemical gradient created by the difference in proton (H+) concentration across the inner mitochondrial membrane in eukaryotes or the plasma membrane in bacteria. During photosynthesis in plants, ATP is synthesized by ATP synthase using a proton gradient created in the thylakoid lumen through the thylakoid membrane and into the chloroplast stro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transmembrane Transporters
A transmembrane protein (TP) is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents. The peptide sequence that spans the membrane, or the transmembrane segment, is largely hydrophobic and can be visualized using the hydropathy plot. Depending on the number of transmembrane segments, transmembrane proteins can be classified as single-span (or bitopic) or multi-span (polytopic). Some other integral membrane proteins are called monotopic, meaning that they are also permanently attached to the membrane, but do not pass t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
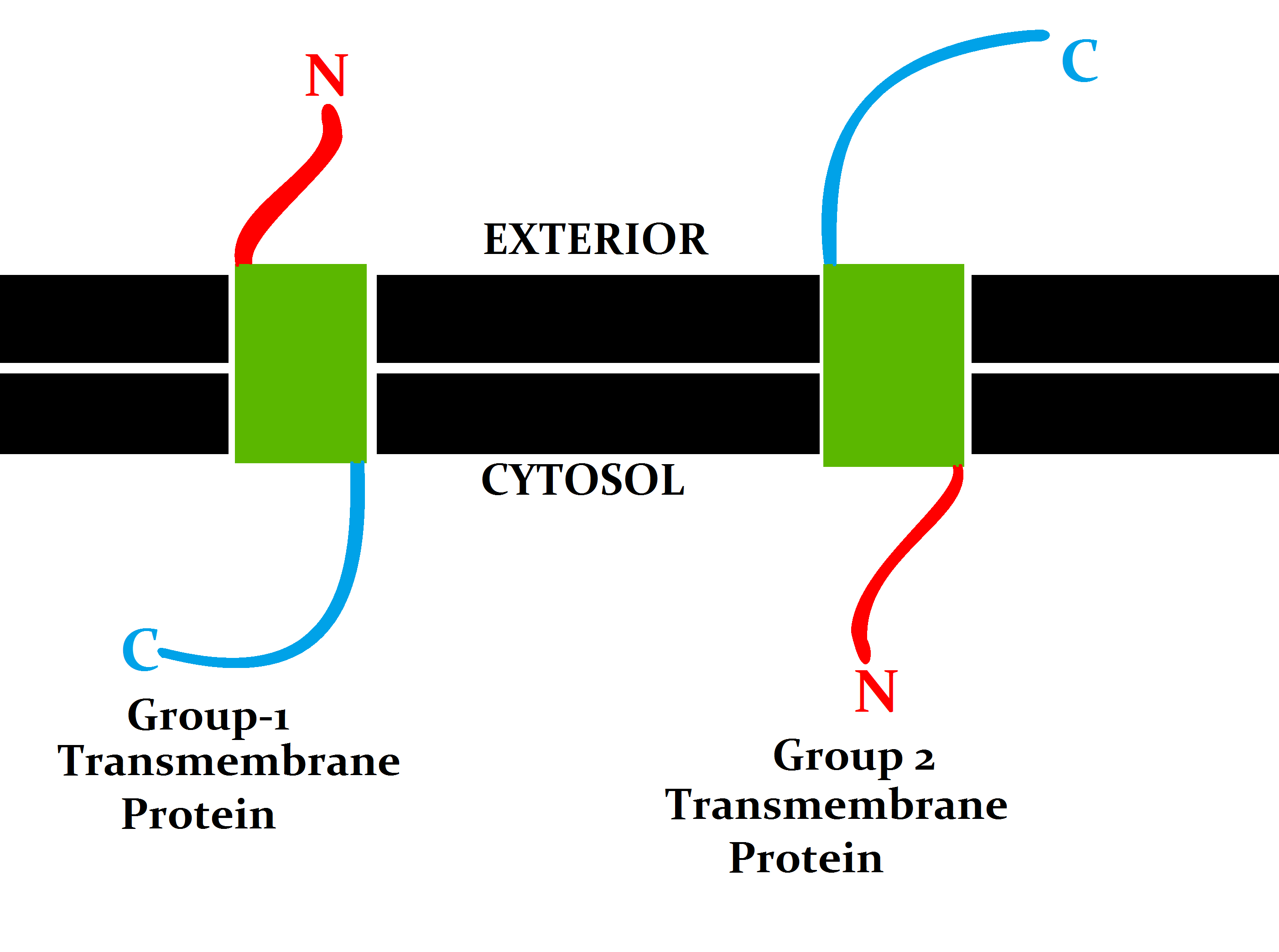
Transmembrane Proteins
A transmembrane protein (TP) is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents. The peptide sequence that spans the membrane, or the transmembrane segment, is largely hydrophobic and can be visualized using the hydropathy plot. Depending on the number of transmembrane segments, transmembrane proteins can be classified as single-span (or bitopic) or multi-span (polytopic). Some other integral membrane proteins are called monotopic, meaning that they are also permanently attached to the membrane, but do not pass t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
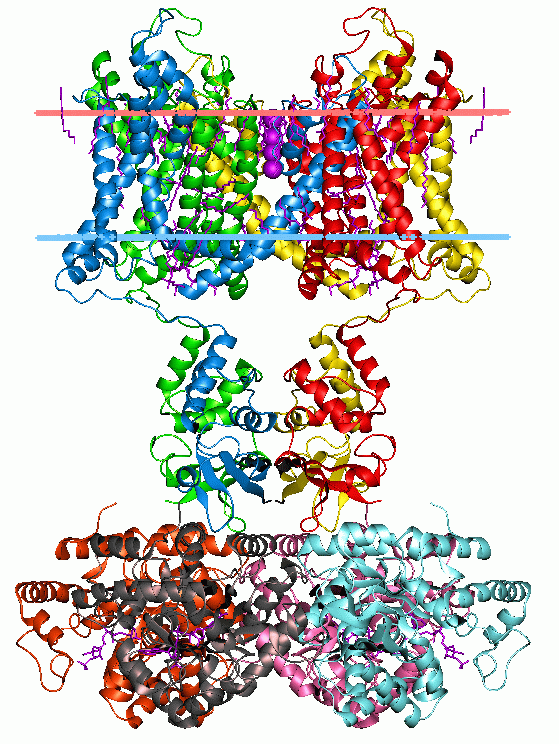
Membrane Proteins
Membrane proteins are common proteins that are part of, or interact with, biological membranes. Membrane proteins fall into several broad categories depending on their location. Integral membrane proteins are a permanent part of a cell membrane and can either penetrate the membrane (transmembrane) or associate with one or the other side of a membrane ( integral monotopic). Peripheral membrane proteins are transiently associated with the cell membrane. Membrane proteins are common, and medically important—about a third of all human proteins are membrane proteins, and these are targets for more than half of all drugs. Nonetheless, compared to other classes of proteins, determining membrane protein structures remains a challenge in large part due to the difficulty in establishing experimental conditions that can preserve the correct conformation of the protein in isolation from its native environment. Function Membrane proteins perform a variety of functions vital to the surv ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Families
A protein family is a group of evolutionarily related proteins. In many cases, a protein family has a corresponding gene family, in which each gene encodes a corresponding protein with a 1:1 relationship. The term "protein family" should not be confused with family as it is used in taxonomy. Proteins in a family descend from a common ancestor and typically have similar three-dimensional structures, functions, and significant sequence similarity. The most important of these is sequence similarity (usually amino-acid sequence), since it is the strictest indicator of homology and therefore the clearest indicator of common ancestry. A fairly well developed framework exists for evaluating the significance of similarity between a group of sequences using sequence alignment methods. Proteins that do not share a common ancestor are very unlikely to show statistically significant sequence similarity, making sequence alignment a powerful tool for identifying the members of protein familie ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquinone Reductase (H+-translocating)
Ubiquinone reductase may refer to: * NADH dehydrogenase NADH dehydrogenase is an enzyme that converts nicotinamide adenine dinucleotide (NAD) from its reduced form (NADH) to its oxidized form (NAD+). Members of the NADH dehydrogenase family and analogues are commonly systematically named using the for ..., an enzyme * NADH:ubiquinone reductase (non-electrogenic), an enzyme {{Short pages monitor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
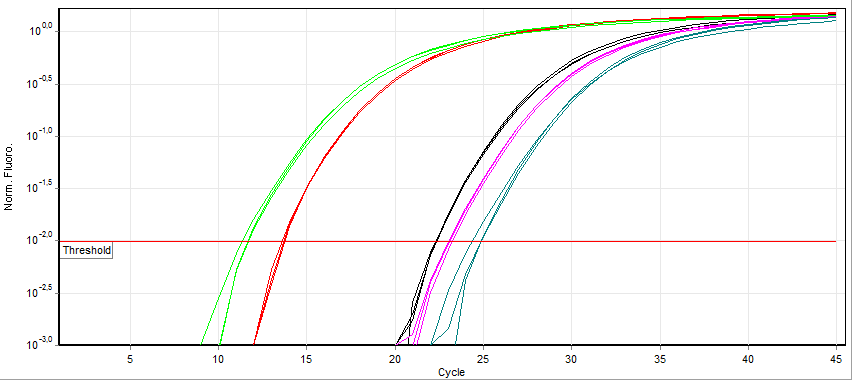
RT-qPCR
A real-time polymerase chain reaction (real-time PCR, or qPCR) is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR (i.e., in real time), not at its end, as in conventional PCR. Real-time PCR can be used quantitatively (quantitative real-time PCR) and semi-quantitatively (i.e., above/below a certain amount of DNA molecules) (semi-quantitative real-time PCR). Two common methods for the detection of PCR products in real-time PCR are (1) non-specific fluorescent dyes that intercalate with any double-stranded DNA and (2) sequence-specific DNA probes consisting of oligonucleotides that are labelled with a fluorescent reporter, which permits detection only after hybridization of the probe with its complementary sequence. The Minimum Information for Publication of Quantitative Real-Time PCR Experiments ( MIQE) guidelines propose that the abbreviation ''qPCR'' be used for ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Archaea
Archaea ( ; singular archaeon ) is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria (in the Archaebacteria kingdom), but this term has fallen out of use. Archaeal cells have unique properties separating them from the other two domains, Bacteria and Eukaryota. Archaea are further divided into multiple recognized phyla. Classification is difficult because most have not been isolated in a laboratory and have been detected only by their gene sequences in environmental samples. Archaea and bacteria are generally similar in size and shape, although a few archaea have very different shapes, such as the flat, square cells of ''Haloquadratum walsbyi''. Despite this morphological similarity to bacteria, archaea possess genes and several metabolic pathways that are more closely related to those of eukaryotes, notably for the enzymes involved ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methanogen
Methanogens are microorganisms that produce methane as a metabolic byproduct in hypoxic conditions. They are prokaryotic and belong to the domain Archaea. All known methanogens are members of the archaeal phylum Euryarchaeota. Methanogens are common in wetlands, where they are responsible for marsh gas, and in the digestive tracts of animals such as ruminants and many humans, where they are responsible for the methane content of belching in ruminants and flatulence in humans. In marine sediments, the biological production of methane, also termed methanogenesis, is generally confined to where sulfates are depleted, below the top layers. Moreover, methanogenic archaea populations play an indispensable role in anaerobic wastewater treatments. Others are extremophiles, found in environments such as hot springs and submarine hydrothermal vents as well as in the "solid" rock of Earth's crust, kilometers below the surface. Physical description Methanogens are coccoid (spherical shap ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cytochrome
Cytochromes are redox-active proteins containing a heme, with a central Fe atom at its core, as a cofactor. They are involved in electron transport chain and redox catalysis. They are classified according to the type of heme and its mode of binding. Four varieties are recognized by the International Union of Biochemistry and Molecular Biology (IUBMB), cytochromes a, cytochromes b, cytochromes c and cytochrome d. Cytochrome function is linked to the reversible redox change from ferrous (Fe(II)) to the ferric (Fe(III)) oxidation state of the iron found in the heme core. In addition to the classification by the IUBMB into four cytochrome classes, several additional classifications such as cytochrome o and cytochrome P450 can be found in biochemical literature. History Cytochromes were initially described in 1884 by Charles Alexander MacMunn as respiratory pigments (myohematin or histohematin). In the 1920s, Keilin rediscovered these respiratory pigments and named them the c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |


.jpg)