|
Evolution@Home
evolution@home was a volunteer computing project for evolutionary biology, launched in 2001. The aim of evolution@home is to improve understanding of evolutionary processes. This is achieved by simulating individual-based models. The Simulator005 module of evolution@home was designed to better predict the behaviour of Muller's ratchet. The project was operated semi-automatically; participants had to manually download tasks from the webpage and submit results by email using this method of operation. yoyo@home used a BOINC wrapper to completely automate this project by automatically distributing tasks and collecting their results. Therefore, the BOINC version was a complete volunteer computing project. yoyo@home has declared its involvement in this project finished. See also * Artificial life *Digital organism *Evolutionary computation In computer science, evolutionary computation is a family of algorithms for global optimization inspired by biological evolution, and th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
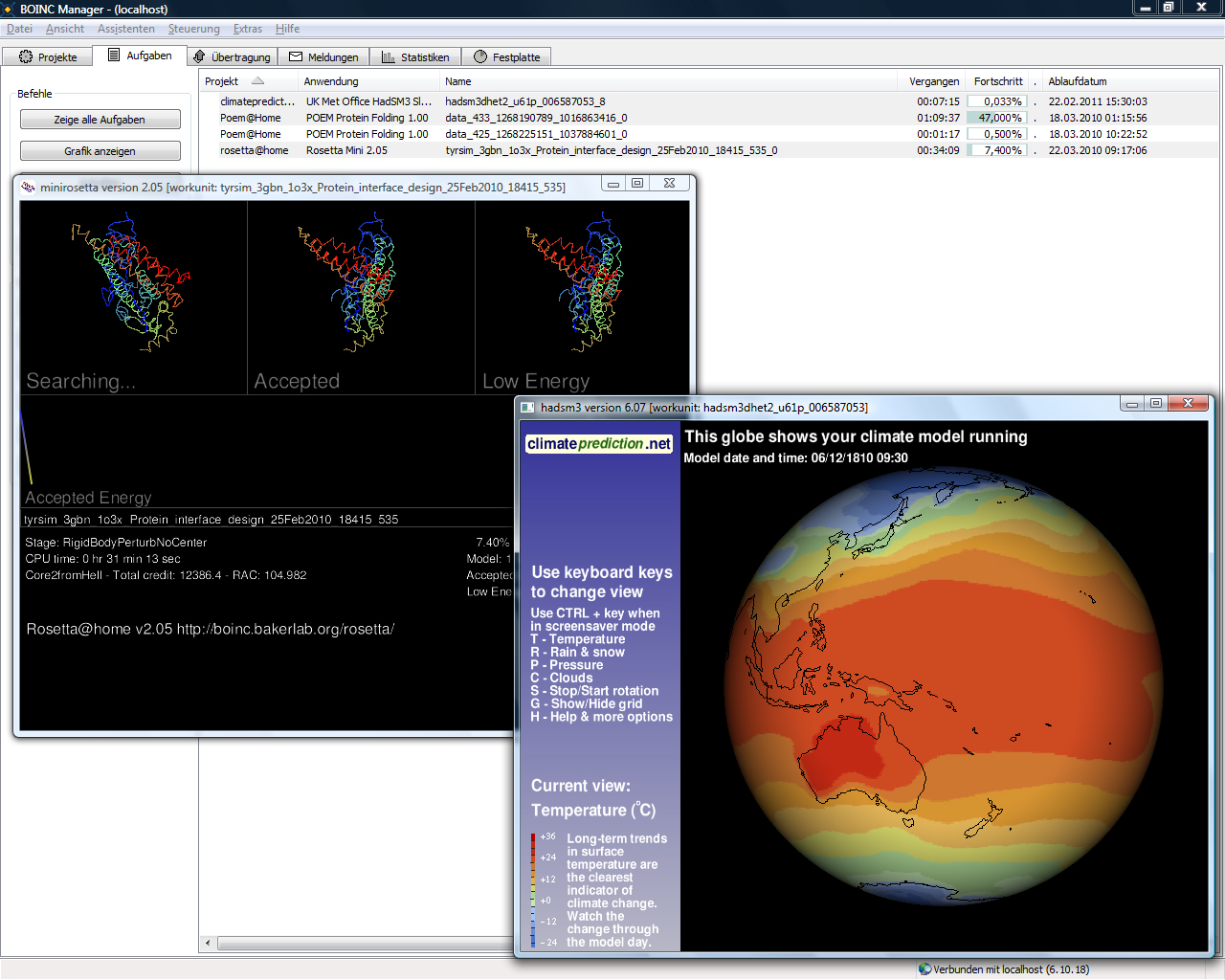
BOINC
The Berkeley Open Infrastructure for Network Computing (BOINC, pronounced – rhymes with "oink") is an open-source middleware system for volunteer computing (a type of distributed computing). Developed originally to support SETI@home, it became the platform for many other applications in areas as diverse as medicine, molecular biology, mathematics, linguistics, climatology, environmental science, and astrophysics, among others. The purpose of BOINC is to enable researchers to utilize processing resources of personal computers and other devices around the world. BOINC development began with a group based at the Space Sciences Laboratory (SSL) at the University of California, Berkeley, and led by David P. Anderson, who also led SETI@home. As a high-performance volunteer computing platform, BOINC brings together 34,236 active participants employing 136,341 active computers (hosts) worldwide, processing daily on average 20.164 PetaFLOPS (it would be the 21st largest processing ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Berkeley Open Infrastructure For Network Computing
The Berkeley Open Infrastructure for Network Computing (BOINC, pronounced – rhymes with "oink") is an open-source middleware system for volunteer computing (a type of distributed computing). Developed originally to support SETI@home, it became the platform for many other applications in areas as diverse as medicine, molecular biology, mathematics, linguistics, climatology, environmental science, and astrophysics, among others. The purpose of BOINC is to enable researchers to utilize processing resources of personal computers and other devices around the world. BOINC development began with a group based at the Space Sciences Laboratory (SSL) at the University of California, Berkeley, and led by David P. Anderson, who also led SETI@home. As a high-performance volunteer computing platform, BOINC brings together 34,236 active participants employing 136,341 active computers (hosts) worldwide, processing daily on average 20.164 PetaFLOPS (it would be the 21st largest processin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Volunteer Computing Projects
This is a comprehensive list of volunteer computing projects; a type of distributed computing where volunteers donate computing time to specific causes. The donated computing power comes from idle CPUs and GPUs in personal computers, video game consoles and Android devices. Each project seeks to utilize the computing power of many internet connected devices to solve problems and perform tedious, repetitive research in a very cost effective manner. Active projects Completed projects See also * List of grid computing projects * List of citizen science projects * List of crowdsourcing projects Below is a list of projects that rely on crowdsourcing. See also open innovation. A *Adaptive Vehicle Make is a project overseen by DARPA to crowdsource the design and manufacture of a new Armored fighting vehicle, armored vehicle. *Air Qual ... * List of free and open-source Android applications * List of Berkeley Open Infrastructure for Network Computing (BOINC) project ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Digital Organism
A digital organism is a self-replicating computer program that mutates and evolves. Digital organisms are used as a tool to study the dynamics of Darwinian evolution, and to test or verify specific hypotheses or mathematical models of evolution. The study of digital organisms is closely related to the area of artificial life. History Digital organisms can be traced back to the game Darwin, developed in 1961 at Bell Labs, in which computer programs had to compete with each other by trying to stop others from executing . A similar implementation that followed this was the game Core War. In Core War, it turned out that one of the winning strategies was to replicate as fast as possible, which deprived the opponent of all computational resources. Programs in the Core War game were also able to mutate themselves and each other by overwriting instructions in the simulated "memory" in which the game took place. This allowed competing programs to embed damaging instructions in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Digital Organisms
A digital organism is a self-replicating computer program that mutates and evolves. Digital organisms are used as a tool to study the dynamics of Darwinian evolution, and to test or verify specific hypotheses or mathematical models of evolution. The study of digital organisms is closely related to the area of artificial life. History Digital organisms can be traced back to the game Darwin, developed in 1961 at Bell Labs, in which computer programs had to compete with each other by trying to stop others from executing . A similar implementation that followed this was the game Core War. In Core War, it turned out that one of the winning strategies was to replicate as fast as possible, which deprived the opponent of all computational resources. Programs in the Core War game were also able to mutate themselves and each other by overwriting instructions in the simulated "memory" in which the game took place. This allowed competing programs to embed damaging instructions ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Muller's Ratchet
In evolutionary genetics, Muller's ratchet (named after Hermann Joseph Muller, by analogy with a ratchet effect) is a process through which, in the absence of recombination (especially in an asexual population), an accumulation of irreversible deleterious mutations results. (original paper as cited by, e.g.: ; ) This happens due to the fact that in the absence of recombination, and assuming reverse mutations are rare, offspring bear at least as much mutational load as their parents. Muller proposed this mechanism as one reason why sexual reproduction may be favored over asexual reproduction, as sexual organisms benefit from recombination and consequent elimination of deleterious mutations. The negative effect of accumulating irreversible deleterious mutations may not be prevalent in organisms which, while they reproduce asexually, also undergo other forms of recombination. This effect has also been observed in those regions of the genomes of sexual organisms that do not underg ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Agent-based Model
An agent-based model (ABM) is a computational model for simulating the actions and interactions of autonomous agents (both individual or collective entities such as organizations or groups) in order to understand the behavior of a system and what governs its outcomes. It combines elements of game theory, complex systems, emergence, computational sociology, multi-agent systems, and evolutionary programming. Monte Carlo methods are used to understand the stochasticity of these models. Particularly within ecology, ABMs are also called individual-based models (IBMs). A review of recent literature on individual-based models, agent-based models, and multiagent systems shows that ABMs are used in many scientific domains including biology, ecology and social science. Agent-based modeling is related to, but distinct from, the concept of multi-agent systems or multi-agent simulation in that the goal of ABM is to search for explanatory insight into the collective behavior of agents obeying ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Volunteer Computing
Volunteer computing is a type of distributed computing in which people donate their computers' unused resources to a research-oriented project, and sometimes in exchange for credit points. The fundamental idea behind it is that a modern desktop computer is sufficiently powerful to perform billions of operations a second, but for most users only between 10-15% of its capacity is used. Typical uses like basic word processing or web browsing leave the computer mostly idle. The practice of volunteer computing, which dates back to the mid-1990s, can potentially make substantial processing power available to researchers at minimal cost. Typically, a program running on a volunteer's computer periodically contacts a research application to request jobs and report results. A middleware system usually serves as an intermediary. History The first volunteer computing project was the Great Internet Mersenne Prime Search, which was started in January 1996. It was followed in 1997 by distribute ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Evolution
Evolution is change in the heritable characteristics of biological populations over successive generations. These characteristics are the expressions of genes, which are passed on from parent to offspring during reproduction. Variation tends to exist within any given population as a result of genetic mutation and recombination. Evolution occurs when evolutionary processes such as natural selection (including sexual selection) and genetic drift act on this variation, resulting in certain characteristics becoming more common or more rare within a population. The evolutionary pressures that determine whether a characteristic is common or rare within a population constantly change, resulting in a change in heritable characteristics arising over successive generations. It is this process of evolution that has given rise to biodiversity at every level of biological organisation, including the levels of species, individual organisms, and molecules. The theory of evol ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Evolutionary Computation
In computer science, evolutionary computation is a family of algorithms for global optimization inspired by biological evolution, and the subfield of artificial intelligence and soft computing studying these algorithms. In technical terms, they are a family of population-based trial and error problem solvers with a metaheuristic or stochastic optimization character. In evolutionary computation, an initial set of candidate solutions is generated and iteratively updated. Each new generation is produced by stochastically removing less desired solutions, and introducing small random changes. In biological terminology, a population of solutions is subjected to natural selection (or artificial selection) and mutation. As a result, the population will gradually evolve to increase in fitness, in this case the chosen fitness function of the algorithm. Evolutionary computation techniques can produce highly optimized solutions in a wide range of problem settings, making them ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Folding@home
Folding@home (FAH or F@h) is a volunteer computing project aimed to help scientists develop new therapeutics for a variety of diseases by the means of simulating protein dynamics. This includes the process of protein folding and the movements of proteins, and is reliant on simulations run on volunteers' personal computers. Folding@home is currently based at the University of Pennsylvania and led by Greg Bowman, a former student of Vijay Pande. The project utilizes graphics processing units (GPUs), central processing units (CPUs), and ARM processors like those on the Raspberry Pi for volunteer computing and scientific research. The project uses statistical simulation methodology that is a paradigm shift from traditional computing methods. As part of the client–server model network architecture, the volunteered machines each receive pieces of a simulation (work units), complete them, and return them to the project's database servers, where the units are compiled into an ov ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |