|
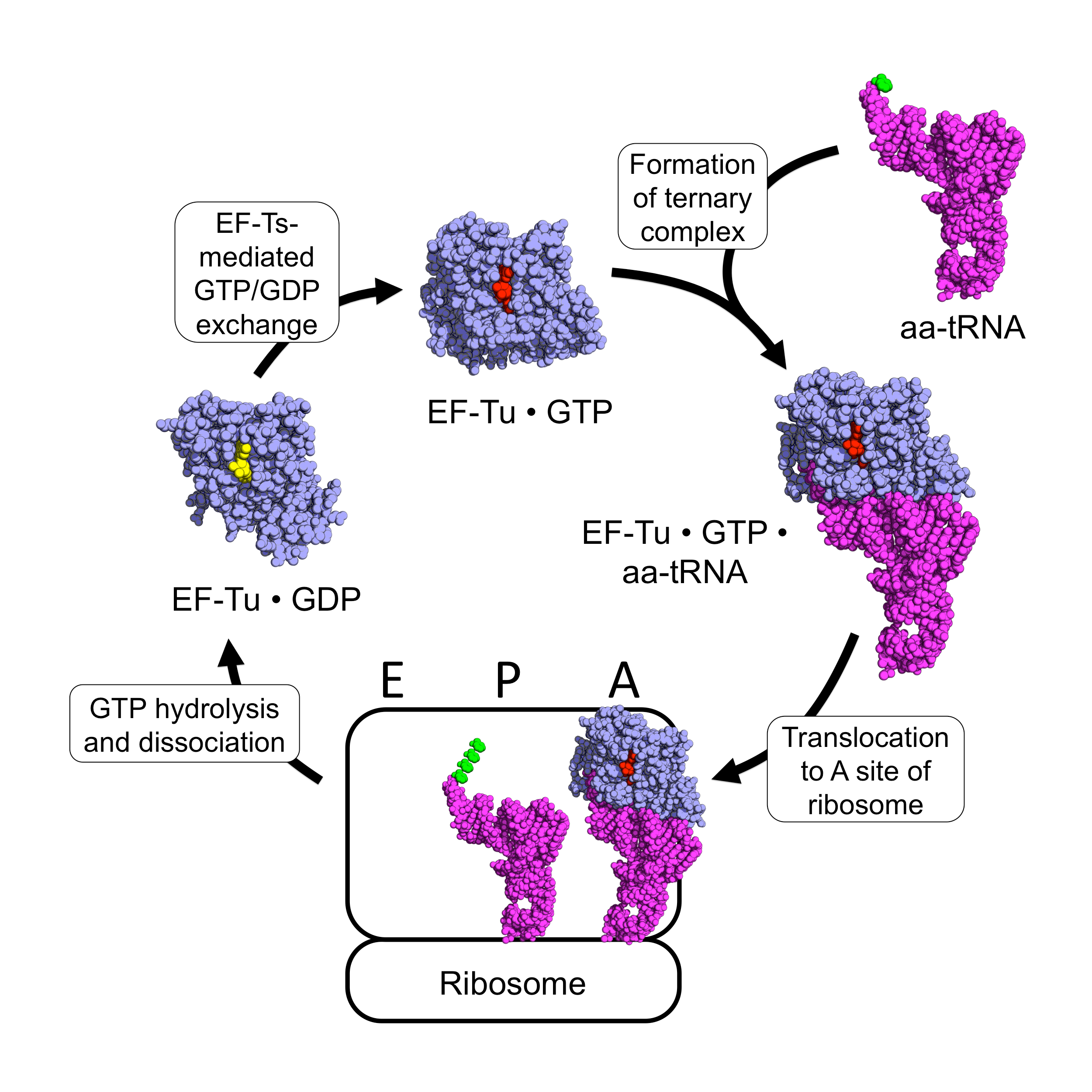
EF-Tu Cycle
EF-Tu (elongation factor thermo unstable) is a prokaryotic elongation factor responsible for catalyzing the binding of an aminoacyl-tRNA (aa-tRNA) to the ribosome. It is a G-protein, and facilitates the selection and binding of an aa-tRNA to the A-site of the ribosome. As a reflection of its crucial role in translation, EF-Tu is one of the most abundant and highly conserved proteins in prokaryotes. It is found in eukaryotic mitochondria as TUFM. As a family of elongation factors, EF-Tu also includes its eukaryotic and archaeal homolog, the alpha subunit of eEF-1 (EF-1A). Background Elongation factors are part of the mechanism that synthesizes new proteins through translation in the ribosome. Transfer RNAs (tRNAs) carry the individual amino acids that become integrated into a protein sequence, and have an anticodon for the specific amino acid that they are charged with. Messenger RNA (mRNA) carries the genetic information that encodes the primary structure of a protein, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Prokaryotic Elongation Factors
Elongation factors are a set of proteins that function at the ribosome, during protein synthesis, to facilitate translational elongation from the formation of the first to the last peptide bond of a growing polypeptide. Most common elongation factors in prokaryotes are EF-Tu, EF-Ts, EF-G. Bacteria and eukaryotes use elongation factors that are largely homologous to each other, but with distinct structures and different research nomenclatures. Elongation is the most rapid step in translation. In bacteria, it proceeds at a rate of 15 to 20 amino acids added per second (about 45-60 nucleotides per second). In eukaryotes the rate is about two amino acids per second (about 6 nucleotides read per second). Elongation factors play a role in orchestrating the events of this process, and in ensuring the high accuracy translation at these speeds. Nomenclature of homologous EFs In addition to their cytoplasmic machinery, eukaryotic mitochondria and plastids have their own translation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EF-Tu Cycle
EF-Tu (elongation factor thermo unstable) is a prokaryotic elongation factor responsible for catalyzing the binding of an aminoacyl-tRNA (aa-tRNA) to the ribosome. It is a G-protein, and facilitates the selection and binding of an aa-tRNA to the A-site of the ribosome. As a reflection of its crucial role in translation, EF-Tu is one of the most abundant and highly conserved proteins in prokaryotes. It is found in eukaryotic mitochondria as TUFM. As a family of elongation factors, EF-Tu also includes its eukaryotic and archaeal homolog, the alpha subunit of eEF-1 (EF-1A). Background Elongation factors are part of the mechanism that synthesizes new proteins through translation in the ribosome. Transfer RNAs (tRNAs) carry the individual amino acids that become integrated into a protein sequence, and have an anticodon for the specific amino acid that they are charged with. Messenger RNA (mRNA) carries the genetic information that encodes the primary structure of a protein, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Karissa Sanbonmatsu
Karissa Y. Sanbonmatsu is an American structural biologist at Los Alamos National Laboratory. She works on the mechanism of non-coding RNA complexes including the ribosome, riboswitches, long non-coding RNAs, as well as chromatin. She was the first to perform an atomistic simulation of the ribosome, determine the secondary structure of an intact lncRNA and to publish one billion atom simulationof a biomolecular complex. Education and early career Sanbonmatsu was born in Rochester, New York, the daughter of Joan Loveridge-Sanbonmatsu, and Akira Loveridge-Sanbonmatsu, who are both professors of speech communication in the State University of New York. She attended Oswego High School, and was valedictorian. She won the Pembroke College Stokes Society Scientific Lecture Competition at the University of Cambridge. Sanbonmatsu studied physics at Columbia University, where she used the Very Large Array radio telescope to estimate the distance to supernova remnant G27.4+0.0 and its ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
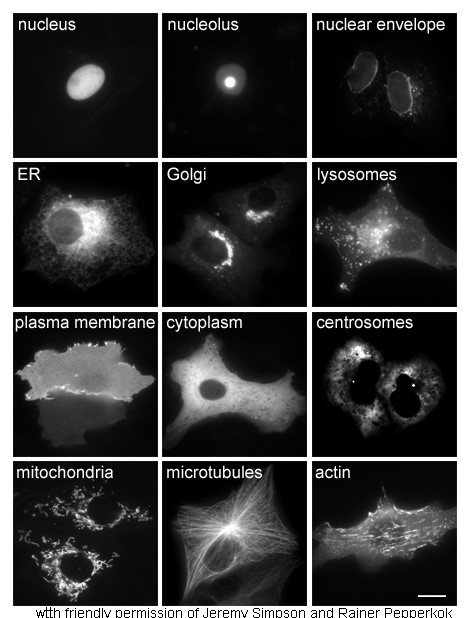
Cytoplasm
In cell biology, the cytoplasm is all of the material within a eukaryotic cell, enclosed by the cell membrane, except for the cell nucleus. The material inside the nucleus and contained within the nuclear membrane is termed the nucleoplasm. The main components of the cytoplasm are cytosol (a gel-like substance), the organelles (the cell's internal sub-structures), and various cytoplasmic inclusions. The cytoplasm is about 80% water and is usually colorless. The submicroscopic ground cell substance or cytoplasmic matrix which remains after exclusion of the cell organelles and particles is groundplasm. It is the hyaloplasm of light microscopy, a highly complex, polyphasic system in which all resolvable cytoplasmic elements are suspended, including the larger organelles such as the ribosomes, mitochondria, the plant plastids, lipid droplets, and vacuoles. Most cellular activities take place within the cytoplasm, such as many metabolic pathways including glycolysis, and proc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EF-Ts
EF-Ts (elongation factor thermo stable) is one of the prokaryotic elongation factors. It is found in human mitochrondria as TSFM. It is similar to eukaryotic EF-1B. EF-Ts serves as the guanine nucleotide exchange factor for EF-Tu (elongation factor thermo unstable), catalyzing the release of guanosine diphosphate from EF-Tu. This enables EF-Tu to bind to a new guanosine triphosphate molecule, release EF-Ts, and go on to catalyze another aminoacyl tRNA addition. Structure The protein Qβ-Replicase is a tetrameric protein, meaning it contains four subunits. These subunits are the two elongation factors, EF-Tu & EF-Ts, the ribosomal protein subunit S1, and the RNA dependent RNA polymerase β-subunit. The two elongation factors form a heterodimer structure known as The Elongation factor complex, which is necessary for the polymerization activity of the RDRP β-Subunit. Its secondary structural components consists of α-helices, β-sheets and β-barrels. EF-Ts comprises the majo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GTPase-activating Protein
GTPase-activating proteins or GTPase-accelerating proteins (GAPs) are a family of regulatory proteins whose members can bind to activated G proteins and stimulate their GTPase activity, with the result of terminating the signaling event. GAPs are also known as RGS protein, or RGS proteins,Kimple, A.J. "Structural Determinants of G-protein α Subunit Selectivity by Regulator of G-protein Signaling 2 (RGS2)". ''The Journal of Biological Chemistry''. 284 (2009): 19402-19411. and these proteins are crucial in controlling the activity of G proteins. Regulation of G proteins is important because these proteins are involved in a variety of important cellular processes. The large G proteins, for example, are involved in transduction of signaling from the G protein-coupled receptor for a variety of signaling processes like hormonal signaling, and small G proteins are involved in processes like cellular trafficking and cell cycling. GAP's role in this function is to turn the G protein's ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphate
In chemistry, a phosphate is an anion, salt, functional group or ester derived from a phosphoric acid. It most commonly means orthophosphate, a derivative of orthophosphoric acid . The phosphate or orthophosphate ion is derived from phosphoric acid by the removal of three protons . Removal of one or two protons gives the dihydrogen phosphate ion and the hydrogen phosphate ion ion, respectively. These names are also used for salts of those anions, such as ammonium dihydrogen phosphate and trisodium phosphate. File:3-phosphoric-acid-3D-balls.png, Phosphoricacid File:2-dihydrogenphosphate-3D-balls.png, Dihydrogenphosphate File:1-hydrogenphosphate-3D-balls.png, Hydrogenphosphate File:0-phosphate-3D-balls.png, Phosphate In organic chemistry, phosphate or orthophosphate is an organophosphate, an ester of orthophosphoric acid of the form where one or more hydrogen atoms are replaced by organic groups. An example is trimethyl phosphate, . The term also refers to the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Guanosine Diphosphate
Guanosine diphosphate, abbreviated GDP, is a nucleoside diphosphate. It is an ester of pyrophosphoric acid with the nucleoside guanosine. GDP consists of a pyrophosphate group, a pentose sugar ribose, and the nucleobase guanine. GDP is the product of GTP dephosphorylation by GTPases, e.g., the G-proteins that are involved in signal transduction. GDP is converted into GTP with the help of pyruvate kinase and phosphoenolpyruvate. See also * DNA *Guanosine triphosphate *Nucleoside *Nucleotide *Oligonucleotide *RNA Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and deoxyribonucleic acid ( DNA) are nucleic acids. Along with lipids, proteins, and carbohydra ... References {{DEFAULTSORT:Guanosine phosphate2 Nucleotides Phosphate esters Purines Pyrophosphates ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrolysis
Hydrolysis (; ) is any chemical reaction in which a molecule of water breaks one or more chemical bonds. The term is used broadly for substitution, elimination, and solvation reactions in which water is the nucleophile. Biological hydrolysis is the cleavage of biomolecules where a water molecule is consumed to effect the separation of a larger molecule into component parts. When a carbohydrate is broken into its component sugar molecules by hydrolysis (e.g., sucrose being broken down into glucose and fructose), this is recognized as saccharification. Hydrolysis reactions can be the reverse of a condensation reaction in which two molecules join into a larger one and eject a water molecule. Thus hydrolysis adds water to break down, whereas condensation builds up by removing water. Types Usually hydrolysis is a chemical process in which a molecule of water is added to a substance. Sometimes this addition causes both the substance and water molecule to split into two parts. I ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GTPase
GTPases are a large family of hydrolase enzymes that bind to the nucleotide guanosine triphosphate (GTP) and hydrolyze it to guanosine diphosphate (GDP). The GTP binding and hydrolysis takes place in the highly conserved P-loop "G domain", a protein domain common to many GTPases. Functions GTPases function as molecular switches or timers in many fundamental cellular processes. Examples of these roles include: * Signal transduction in response to activation of cell surface receptors, including transmembrane receptors such as those mediating taste, smell and vision. * Protein biosynthesis (a.k.a. translation) at the ribosome. * Regulation of cell differentiation, proliferation, division and movement. * Translocation of proteins through membranes. * Transport of vesicles within the cell, and vesicle-mediated secretion and uptake, through GTPase control of vesicle coat assembly. GTPases are active when bound to GTP and inactive when bound to GDP. In the generalized r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Side Chain
In organic chemistry and biochemistry, a side chain is a chemical group that is attached to a core part of the molecule called the "main chain" or backbone. The side chain is a hydrocarbon branching element of a molecule that is attached to a larger hydrocarbon backbone. It is one factor in determining a molecule's properties and reactivity. A side chain is also known as a pendant chain, but a pendant group (side group) has a different definition. Conventions The placeholder R is often used as a generic placeholder for alkyl (saturated hydrocarbon) group side chains in chemical structure diagrams. To indicate other non-carbon groups in structure diagrams, X, Y, or Z are often used. History The ''R'' symbol was introduced by 19th-century French chemist Charles Frédéric Gerhardt, who advocated its adoption on the grounds that it would be widely recognizable and intelligible given its correspondence in multiple European languages to the initial letter of "root" or "residue": ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Selenocysteine
Selenocysteine (symbol Sec or U, in older publications also as Se-Cys) is the 21st proteinogenic amino acid. Selenoproteins contain selenocysteine residues. Selenocysteine is an analogue of the more common cysteine with selenium in place of the sulfur. Selenocysteine is present in several enzymes (for example glutathione peroxidases, tetraiodothyronine 5′ deiodinases, thioredoxin reductases, formate dehydrogenases, glycine reductases, selenophosphate synthetase 2, methionine-''R''-sulfoxide reductase B1 ( SEPX1), and some hydrogenases). It occurs in all three domains of life, including important enzymes (listed above) present in humans. Selenocysteine was discovered by biochemist Thressa Stadtman at the National Institutes of Health. Chemistry Selenocysteine is the Se-analogue of cysteine. It is rarely encountered outside of living tissue (and is not available commercially) because it is very susceptible to air-oxidation. More common is the oxidized derivative sel ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |