EF-Ts on:
[Wikipedia]
[Google]
[Amazon]
EF-Ts (elongation factor thermo stable) is one of the
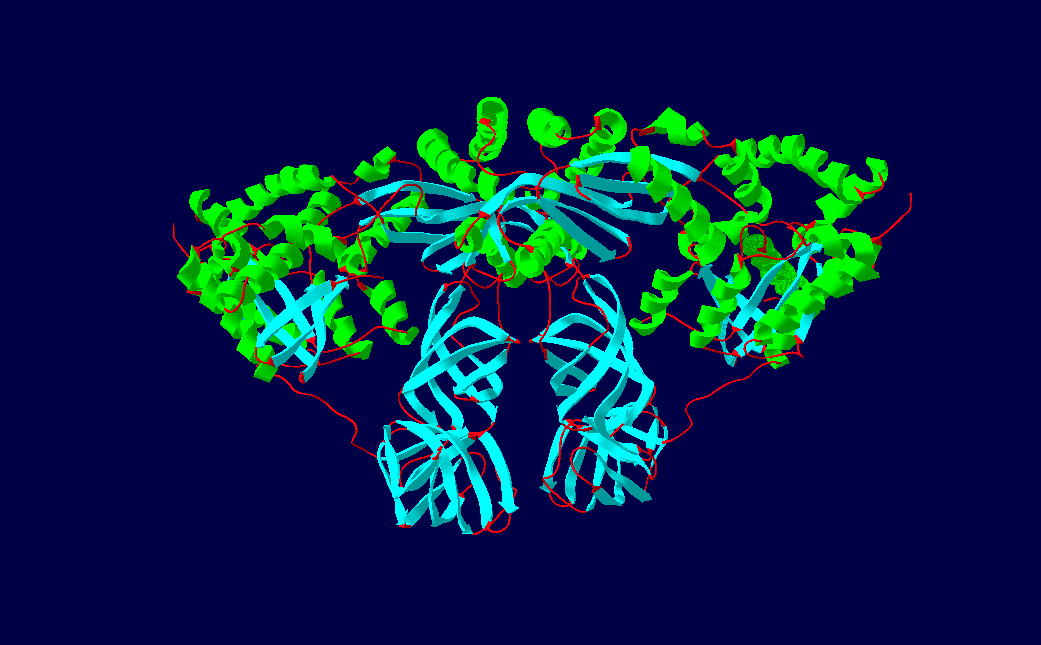 The protein Qβ-Replicase is a tetrameric protein, meaning it contains four subunits. These subunits are the two elongation factors, EF-Tu & EF-Ts, the ribosomal protein subunit S1, and the RNA dependent RNA polymerase β-subunit. The two elongation factors form a heterodimer structure known as The Elongation factor complex, which is necessary for the polymerization activity of the RDRP β-Subunit. Its secondary structural components consists of α-helices, β-sheets and β-barrels.
EF-Ts comprises the majority of the top portion of the protein, while EF-Tu makes up the bottom half where the beta barrels are seen. The conformation is considered to be open, when no guanine nucleotide is bound to the active site in EF-Tu. The EF-Ts chain contains four important domains, the C-terminal domain, N-terminal domain, Dimerization domain, and the Core domain which all play a specific role in the protein's structure and functionality. The dimerization domain contains four anti-parallel α-helices which is the main source of contact between EF-Tu and EF-Ts to form the dimer structure
The protein Qβ-Replicase is a tetrameric protein, meaning it contains four subunits. These subunits are the two elongation factors, EF-Tu & EF-Ts, the ribosomal protein subunit S1, and the RNA dependent RNA polymerase β-subunit. The two elongation factors form a heterodimer structure known as The Elongation factor complex, which is necessary for the polymerization activity of the RDRP β-Subunit. Its secondary structural components consists of α-helices, β-sheets and β-barrels.
EF-Ts comprises the majority of the top portion of the protein, while EF-Tu makes up the bottom half where the beta barrels are seen. The conformation is considered to be open, when no guanine nucleotide is bound to the active site in EF-Tu. The EF-Ts chain contains four important domains, the C-terminal domain, N-terminal domain, Dimerization domain, and the Core domain which all play a specific role in the protein's structure and functionality. The dimerization domain contains four anti-parallel α-helices which is the main source of contact between EF-Tu and EF-Ts to form the dimer structure
 The N-terminal domain spans from resides 1-54 (n1-n54), Core domain is from n55-n179, the Dimerization domain is from n180-n228, and lastly the C-terminal domain that is from n264-n282. the core domain contains two subdomains, C and N which interact with domains 3 and 1 of EF-Tu respectively.
The N-terminal domain spans from resides 1-54 (n1-n54), Core domain is from n55-n179, the Dimerization domain is from n180-n228, and lastly the C-terminal domain that is from n264-n282. the core domain contains two subdomains, C and N which interact with domains 3 and 1 of EF-Tu respectively.
prokaryotic elongation factors
Elongation factors are a set of proteins that function at the ribosome, during protein synthesis, to facilitate translational elongation from the formation of the first to the last peptide bond of a growing polypeptide. Most common elongation f ...
. It is found in human mitochrondria as TSFM
Elongation factor Ts, mitochondrial is a protein that in humans is encoded by the ''TSFM'' gene. It is an EF-Ts
EF-Ts (elongation factor thermo stable) is one of the prokaryotic elongation factors. It is found in human mitochrondria as TSFM. ...
. It is similar to eukaryotic EF-1B.
EF-Ts serves as the guanine
Guanine () ( symbol G or Gua) is one of the four main nucleobases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine (uracil in RNA). In DNA, guanine is paired with cytosine. The guanine nucleoside is called ...
nucleotide exchange factor for EF-Tu
EF-Tu (elongation factor thermo unstable) is a prokaryotic elongation factor responsible for catalyzing the binding of an aminoacyl-tRNA (aa-tRNA) to the ribosome. It is a G-protein, and facilitates the selection and binding of an aa-tRNA to th ...
(elongation factor thermo unstable), catalyzing the release of guanosine diphosphate
Guanosine diphosphate, abbreviated GDP, is a nucleoside diphosphate. It is an ester of pyrophosphoric acid with the nucleoside guanosine. GDP consists of a pyrophosphate group, a pentose sugar ribose, and the nucleobase guanine.
GDP is the product ...
from EF-Tu. This enables EF-Tu to bind to a new guanosine triphosphate
Guanosine-5'-triphosphate (GTP) is a purine nucleoside triphosphate. It is one of the building blocks needed for the synthesis of RNA during the transcription process. Its structure is similar to that of the guanosine nucleoside, the only diffe ...
molecule, release EF-Ts, and go on to catalyze another aminoacyl tRNA
Transfer RNA (abbreviated tRNA and formerly referred to as sRNA, for soluble RNA) is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes), that serves as the physical link between the mRNA and the amino ac ...
addition.
Structure
 The protein Qβ-Replicase is a tetrameric protein, meaning it contains four subunits. These subunits are the two elongation factors, EF-Tu & EF-Ts, the ribosomal protein subunit S1, and the RNA dependent RNA polymerase β-subunit. The two elongation factors form a heterodimer structure known as The Elongation factor complex, which is necessary for the polymerization activity of the RDRP β-Subunit. Its secondary structural components consists of α-helices, β-sheets and β-barrels.
EF-Ts comprises the majority of the top portion of the protein, while EF-Tu makes up the bottom half where the beta barrels are seen. The conformation is considered to be open, when no guanine nucleotide is bound to the active site in EF-Tu. The EF-Ts chain contains four important domains, the C-terminal domain, N-terminal domain, Dimerization domain, and the Core domain which all play a specific role in the protein's structure and functionality. The dimerization domain contains four anti-parallel α-helices which is the main source of contact between EF-Tu and EF-Ts to form the dimer structure
The protein Qβ-Replicase is a tetrameric protein, meaning it contains four subunits. These subunits are the two elongation factors, EF-Tu & EF-Ts, the ribosomal protein subunit S1, and the RNA dependent RNA polymerase β-subunit. The two elongation factors form a heterodimer structure known as The Elongation factor complex, which is necessary for the polymerization activity of the RDRP β-Subunit. Its secondary structural components consists of α-helices, β-sheets and β-barrels.
EF-Ts comprises the majority of the top portion of the protein, while EF-Tu makes up the bottom half where the beta barrels are seen. The conformation is considered to be open, when no guanine nucleotide is bound to the active site in EF-Tu. The EF-Ts chain contains four important domains, the C-terminal domain, N-terminal domain, Dimerization domain, and the Core domain which all play a specific role in the protein's structure and functionality. The dimerization domain contains four anti-parallel α-helices which is the main source of contact between EF-Tu and EF-Ts to form the dimer structure
Domains
 The N-terminal domain spans from resides 1-54 (n1-n54), Core domain is from n55-n179, the Dimerization domain is from n180-n228, and lastly the C-terminal domain that is from n264-n282. the core domain contains two subdomains, C and N which interact with domains 3 and 1 of EF-Tu respectively.
The N-terminal domain spans from resides 1-54 (n1-n54), Core domain is from n55-n179, the Dimerization domain is from n180-n228, and lastly the C-terminal domain that is from n264-n282. the core domain contains two subdomains, C and N which interact with domains 3 and 1 of EF-Tu respectively.
Elongation process pathway
EF-Ts functions as guanine nucleotide exchange factor, it catalyzes the reaction of EF-Tu*GDP ( inactive form) to EF-Tu*GTP (active). EF-Tu (active) then delivers the aminoacyl-tRNA to the ribosome. Therefore, EF-Ts main role is recycling EF-Tu back to its active state in order to complete another elongation cycle. The majority of this pathway is performed through conformational changes of EF-Tu domain 1 which contains the active site and manipulation of the switch 1 & 2 regions by the ribosome and tRNA. First, in domain 1 of EF-Tu the GTPase activity site is blocked by a series of hydrophobic residues that block the catalytic residue His 84 in the inactive form prior to activation via EF-Ts. Once the tRNA is bound to EF-Tu, it is then delivered to the Ribosome which hydrolyses the GTP leaving EF-Tu with a lower affinity to bind the tRNA. The ribosome does this through manipulation of the switch 1 region, after GTP hydrolysis the secondary structure switches from primarily α-helices to β-hairpin. EF-Tu is then released from the ribosome in the inactive state completing the cycle until activated once again by EF-Ts. Helix D of EF-Tu must interact with the N-terminal domain of EF-Ts for guanine nucleotide exchange. A recent study researched the reaction kinetics of the guanine nucleotide exchange by mutating certain residues on helix D of EF-Tu in order to see the primary residues involved in the pathway. Mutation of Leu148 and Glu 152 decreased the rate at which EF-Ts N-terminal domain binds to Helix D significantly, concluding these two residues play an important role in the reaction pathway.Amino acid conservation between organisms
This article focuses on EF-Ts as it exists in Qβ-Bacteriophage, however many organisms use a similar elongation process with proteins that have nearly the same function as EF-Ts. EF-Ts belongs to the group of proteins known as guanine nucleotide exchange factors and these proteins function in many different biochemical pathways, it also belongs to the tsf superfamily. Majority of the amino acid conservation seen between other organisms is located in the N-terminal domain where EF-Ts bind to EF-Tu and the guanine nucleotide exchange occurs. below is the alignment of the important N-Terminal domain of EF-Ts as it exists in other organisms. * ''E.Coli: 8-LVKELRERTGAGMMDCKKALT-20'' * ''LacBS: 8-LVAELRKRTEVSITKAREALS-20'' * ''Bos Taurus:8-LLMKLRRKTGYSFINCKKALE-20'' * ''Drosophila:8-ALAALRKKTGYTFANCKKALE-20'' Conserved amino acids in all four are Leu12 and Arg18 (letters seen in bold above), it can be concluded that these two residues play an important role in the guanine nucleotide exchange since they are the only two completely conserved. In Eukaryotes EF-1 performs the same function, and the mechanism for guanine nucleotide exchange is nearly identical, as EF-Ts but is structurally dissimilar.See also
*Prokaryotic elongation factors
Elongation factors are a set of proteins that function at the ribosome, during protein synthesis, to facilitate translational elongation from the formation of the first to the last peptide bond of a growing polypeptide. Most common elongation f ...
* EF-Tu
EF-Tu (elongation factor thermo unstable) is a prokaryotic elongation factor responsible for catalyzing the binding of an aminoacyl-tRNA (aa-tRNA) to the ribosome. It is a G-protein, and facilitates the selection and binding of an aa-tRNA to th ...
(elongation factor thermo unstable)
* EF-G
EF-G (elongation factor G, historically known as translocase) is a prokaryotic elongation factor involved in protein translation. As a GTPase, EF-G catalyzes the movement (translocation) of transfer RNA (tRNA) and messenger RNA (mRNA) through t ...
(elongation factor G)
* EF-P (elongation factor P)
* Protein translation
In molecular biology and genetics, translation is the process in which ribosomes in the cytoplasm or endoplasmic reticulum synthesize proteins after the process of transcription of DNA to RNA in the cell's nucleus. The entire process is ...
* GTPase
GTPases are a large family of hydrolase enzymes that bind to the nucleotide guanosine triphosphate (GTP) and hydrolyze it to guanosine diphosphate (GDP). The GTP binding and hydrolysis takes place in the highly conserved P-loop "G domain", a pro ...
References
Protein biosynthesis {{enzyme-stub