|
Cross-linking Immunoprecipitation
Cross-linking immunoprecipitation (CLIP) is a method used in molecular biology that combines UV cross-linking with immunoprecipitation in order to analyse protein interactions with RNA or to precisely locate RNA modifications (e.g. m6A). CLIP-based techniques can be used to map RNA binding protein binding sites or RNA modification sites of interest on a genome-wide scale, thereby increasing the understanding of post-transcriptional regulatory networks. Workflow CLIP begins with the in-vivo cross-linking of RNA-protein complexes using ultraviolet light (UV). Upon UV exposure, covalent bonds are formed between proteins and nucleic acids that are in close proximity. The cross-linked cells are then lysed, and the protein of interest is isolated via immunoprecipitation. In order to allow for sequence specific priming of reverse transcription, RNA adapters are ligated to the 3' ends, while radiolabeled phosphates are transferred to the 5' ends of the RNA fragments. The RNA-protein comple ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Biology
Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions. The study of chemical and physical structure of biological macromolecules is known as molecular biology. Molecular biology was first described as an approach focused on the underpinnings of biological phenomena - uncovering the structures of biological molecules as well as their interactions, and how these interactions explain observations of classical biology. In 1945 the term molecular biology was used by physicist William Astbury. In 1953 Francis Crick, James Watson, Rosalind Franklin, and colleagues, working at Medical Research Council unit, Cavendish laboratory, Cambridge (now the MRC Laboratory of Molecular Biology), made a double helix model of DNA which changed the entire research scenario. They proposed the DNA structure based on previous research done by Ro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Degradome Sequencing
Degradome sequencing (Degradome-Seq), also referred to as parallel analysis of RNA ends (PARE), is a modified version of 5'-Rapid Amplification of cDNA Ends (RACE) using high-throughput, deep sequencing methods such aIllumina's SBS technology The degradome encompasses the entire set of proteases that are expressed at a specific time in a given biological material, including tissues, cells, organisms, and biofluids. Thus, sequencing this degradome offers a method for studying and researching the process of RNA degradation. This process is used to identify and quantify RNA degradation products, or fragments, present in any given biological sample. This approach allows for the systematic identification of targets of RNA decay and provides insight into the dynamics of transcriptional and post-transcriptional gene regulation. Degradome sequencing is a complex process which includes multiple steps such as isolating RNA fragments in a given sample as well as ligation and reverse transcrip ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CeRNA
Cerna may refer to: Populated places * Cerna, Croatia, Vukovar-Syrmia County, Croatia * Černá (Žďár nad Sázavou District), Czech Republic * Černá, Semily District, Czech Republic * Cerna, Tulcea, Romania * A village in Vaideeni Commune, Vâlcea County, Romania Rivers Romania * Cerna (Mureș), a tributary of the Mureș in Hunedoara County * Cerna (Danube), a tributary of the Danube in southwestern Romania * Cerna (Olteț), a tributary of the Olteț in Vâlcea County * Cerna (Tulcea), a small tributary of the Danube in Tulcea County * Cerna (Crasna), a tributary of the Crasna in Maramureș and Satu Mare Counties * A tributary of the Mag river in Sibiu County Other rivers * Černá (river), a river in the Czech Republic and Germany People * Cerna (surname) * Černá (surname) Other * ceRNA, competing endogenous RNA, a function of microRNA * Cerna (political organization), part of Anova-Nationalist Brotherhood See also * Černá (other) * Černá Hora (disambigu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
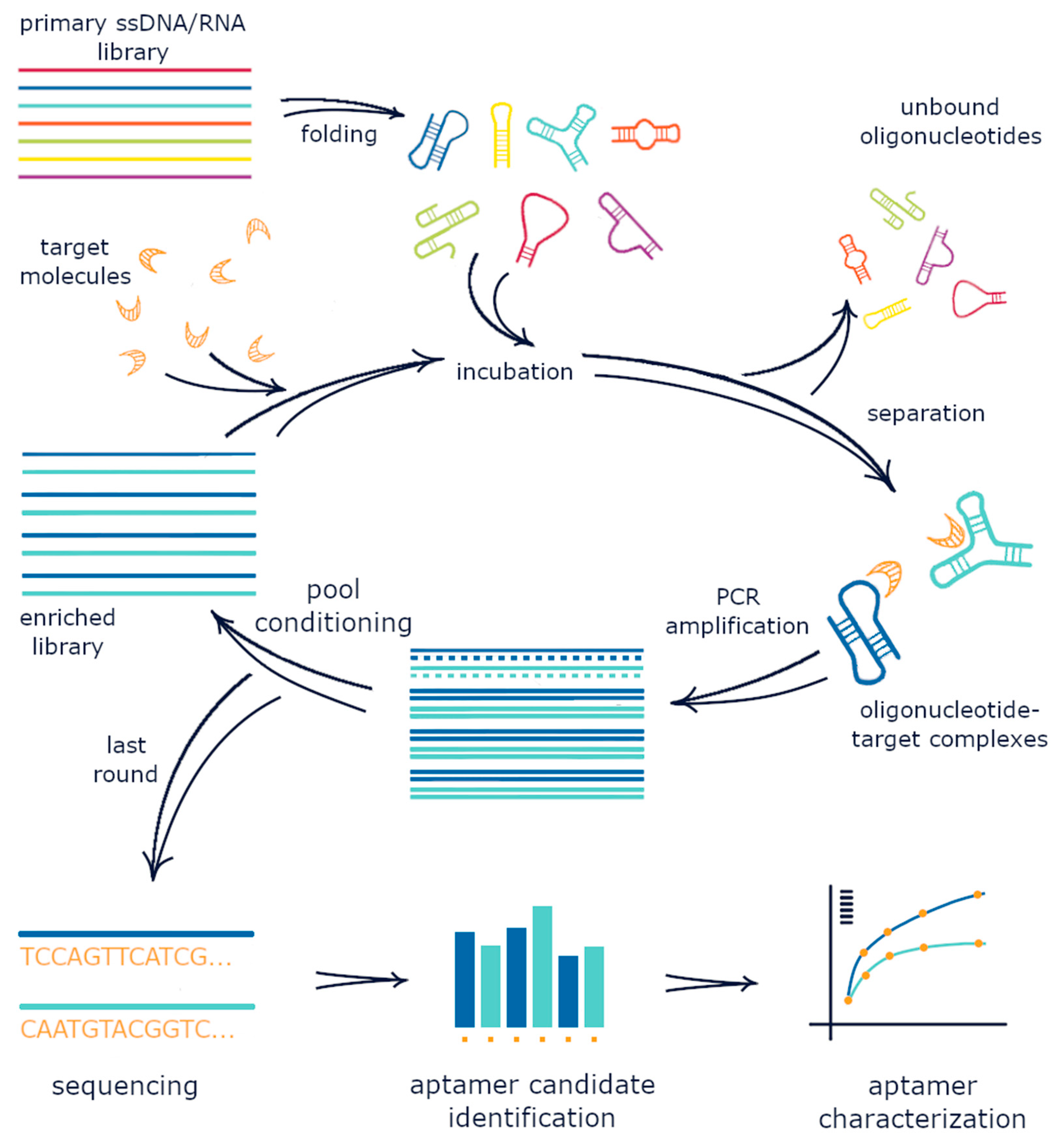
Systematic Evolution Of Ligands By Exponential Enrichment
Systematic evolution of ligands by exponential enrichment (SELEX), also referred to as '' in vitro selection'' or '' in vitro evolution'', is a combinatorial chemistry technique in molecular biology for producing oligonucleotides of either single-stranded DNA or RNA that specifically bind to a target ligand or ligands. These single-stranded DNA or RNA are commonly referred to as aptamers. Although SELEX has emerged as the most commonly used name for the procedure, some researchers have referred to it as SAAB (selected and amplified binding site) and CASTing (cyclic amplification and selection of targets) SELEX was first introduced in 1990. In 2015, a special issue was published in the Journal of Molecular Evolution in the honor of quarter century of the discovery of SELEX. The process begins with the synthesis of a very large oligonucleotide library, consisting of randomly generated sequences of fixed length flanked by constant 5' and 3' ends. The constant ends serve as prim ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ChIP-Seq
ChIP-sequencing, also known as ChIP-seq, is a method used to analyze protein interactions with DNA. ChIP-seq combines chromatin immunoprecipitation (ChIP) with Massively parallel signature sequencing, massively parallel DNA sequencing to identify the binding sites of DNA-associated proteins. It can be used to map global binding sites precisely for any protein of interest. Previously, ChIP-on-chip was the most common technique utilized to study these protein–DNA relations. Uses ChIP-seq is primarily used to determine how transcription factors and other chromatin-associated proteins influence phenotype-affecting mechanisms. Determining how proteins interact with DNA to regulate gene expression is essential for fully understanding many biological processes and disease states. This epigenetic information is complementary to genotype and expression analysis. ChIP-seq technology is currently seen primarily as an alternative to ChIP-chip which requires a protein microarray, hybridization ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RIP-Chip
RIP-chip (RNA immunoprecipitation chip) is a molecular biology technique which combines RNA immunoprecipitation with a microarray. The purpose of this technique is to identify which RNA sequences interact with a particular RNA binding protein of interest in vivo. It can also be used to determine relative levels of gene expression, to identify subsets of RNAs which may be co-regulated, or to identify RNAs that may have related functions. This technique provides insight into the post-transcriptional gene regulation which occurs between RNA and RNA binding proteins. Procedural Overview # Collect and lyse the cells of interest. # Isolate all RNA fragments and the proteins bound to them from the solution. # Immunoprecipitate the protein of interest. The solution containing the protein-bound RNAs is washed over beads which have been conjugated to antibodies. These antibodies are designed to bind to the protein of interest. They pull the protein (and any RNA fragments that are specifi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TargetScan
In bioinformatics, TargetScan is a web server that predicts biological targets of microRNAs (miRNAs) by searching for the presence of sites that match the seed region of each miRNA. For many species, other types of sites, known as 3'-compensatory sites are also identified. These miRNA target predictions are regularly updated and improved by the laboratory of David Bartel in conjunction with the Whitehead Institute Bioinformatics and Research Computing Group. TargetScan includes TargetScanHuman, TargetScanMouse, TargetScanFish, TargetScanFly, and TargetScanWorm. which provide predictions for mammals, zebrafish, insects, and nematodes centered on the genes of human, mouse, zebrafish, ''Drosophila melanogaster'', and ''Caenorhabditis elegans'', respectively. Compared to other target-prediction tools TargetScan provides accurate rankings of the predicted targets for each miRNA. These rankings are based on either the probability of evolutionarily conserved targeting (PCT scores.) or ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
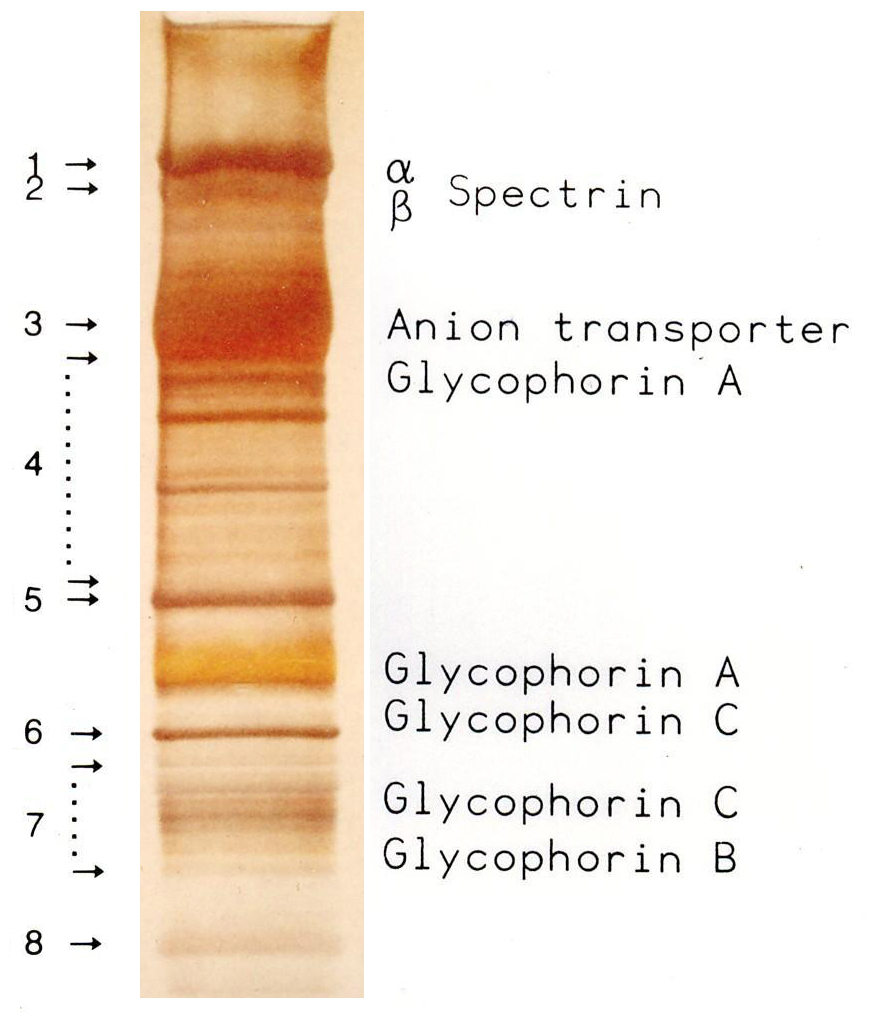
SDS-PAGE
SDS-PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis) is a discontinuous electrophoretic system developed by Ulrich K. Laemmli which is commonly used as a method to separate proteins with molecular masses between 5 and 250 kDa. The combined use of sodium dodecyl sulfate (SDS, also known as sodium lauryl sulfate) and polyacrylamide gel allows to eliminate the influence of structure and charge, and proteins are separated solely on the basis of differences in their molecular weight. Properties SDS-PAGE is an electrophoresis method that allows protein separation by mass. The medium (also referred to as ′matrix′) is a polyacrylamide-based discontinuous gel. The polyacrylamide-gel is typically sandwiched between two glass plates in a slab gel. Although tube gels (in glass cylinders) were used historically, they were rapidly made obsolete with the invention of the more convenient slab gels. In addition, SDS (sodium dodecyl sulfate) is used. About 1.4 grams of S ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
UV Light
Ultraviolet (UV) is a form of electromagnetic radiation with wavelength from 10 nm (with a corresponding frequency around 30 PHz) to 400 nm (750 THz), shorter than that of visible light, but longer than X-rays. UV radiation is present in sunlight, and constitutes about 10% of the total electromagnetic radiation output from the Sun. It is also produced by electric arcs and specialized lights, such as mercury-vapor lamps, tanning lamps, and black lights. Although long-wavelength ultraviolet is not considered an ionizing radiation because its photons lack the energy to ionize atoms, it can cause chemical reactions and causes many substances to glow or fluoresce. Consequently, the chemical and biological effects of UV are greater than simple heating effects, and many practical applications of UV radiation derive from its interactions with organic molecules. Short-wave ultraviolet light damages DNA and sterilizes surfaces with which it comes into contact. For huma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ICLIP
iCLIP (individual-nucleotide resolution CrossLinking and ImmunoPrecipitation) is a variant of the original CLIP method used for identifying protein-RNA interactions, which uses UV light to covalently bind proteins and RNA molecules to identify RNA binding sites of proteins. This crosslinking step has generally less background than standard RNA immunoprecipitation (RIP) protocols, because the covalent bond formed by UV light allows RNA to be fragmented, followed by stringent purification, and this also enables CLIP to identify the positions of protein-RNA interactions. As with all CLIP methods, iCLIP allows for a very stringent purification of the linked protein-RNA complexes by stringent washing during immunoprecipitation followed by SDS-PAGE and transfer to nitrocellulose. The labelled protein-RNA complexes are then visualised for quality control, excised from nitrocellulose, and treated with proteinase to release the RNA, leaving only a few amino acids at the crosslink site of th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ICLIP
iCLIP (individual-nucleotide resolution CrossLinking and ImmunoPrecipitation) is a variant of the original CLIP method used for identifying protein-RNA interactions, which uses UV light to covalently bind proteins and RNA molecules to identify RNA binding sites of proteins. This crosslinking step has generally less background than standard RNA immunoprecipitation (RIP) protocols, because the covalent bond formed by UV light allows RNA to be fragmented, followed by stringent purification, and this also enables CLIP to identify the positions of protein-RNA interactions. As with all CLIP methods, iCLIP allows for a very stringent purification of the linked protein-RNA complexes by stringent washing during immunoprecipitation followed by SDS-PAGE and transfer to nitrocellulose. The labelled protein-RNA complexes are then visualised for quality control, excised from nitrocellulose, and treated with proteinase to release the RNA, leaving only a few amino acids at the crosslink site of th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |