Degradome Sequencing on:
[Wikipedia]
[Google]
[Amazon]
Degradome sequencing (Degradome-Seq), also referred to as parallel analysis of RNA ends (PARE), is a modified version of 5'-
Illumina's SBS technology
The degradome encompasses the entire set of

 The applications of degradome sequencing include identifying
The applications of degradome sequencing include identifying
 In this study, researchers tracked and reported miRNA processing intermediates. Degradome signals on miRNA precursors were extracted and processed for 15 different species. The use of degradome sequencing in this study allowed for the collection of data that supported the analysis and processing of many miRNA precursors, with a greater ratio of high-confidence miRNAs annotated in
In this study, researchers tracked and reported miRNA processing intermediates. Degradome signals on miRNA precursors were extracted and processed for 15 different species. The use of degradome sequencing in this study allowed for the collection of data that supported the analysis and processing of many miRNA precursors, with a greater ratio of high-confidence miRNAs annotated in
starBase database
a database for exploring microRNA cleavage sites from degradome sequencing (Degradome-Seq) data.

Rapid Amplification of cDNA Ends
Rapid amplification of cDNA ends (RACE) is a technique used in molecular biology to obtain the full length sequence of an RNA transcript found within a cell. RACE results in the production of a cDNA copy of the RNA sequence of interest, produced th ...
(RACE) using high-throughput, deep sequencing methods such aIllumina's SBS technology
The degradome encompasses the entire set of
protease
A protease (also called a peptidase, proteinase, or proteolytic enzyme) is an enzyme that catalyzes (increases reaction rate or "speeds up") proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the ...
s that are expressed at a specific time in a given biological material, including tissues, cells, organisms, and biofluids. Thus, sequencing this degradome offers a method for studying and researching the process of RNA degradation. This process is used to identify and quantify RNA degradation products, or fragments, present in any given biological sample. This approach allows for the systematic identification of targets of RNA decay and provides insight into the dynamics of transcriptional
Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules calle ...
and post-transcriptional gene regulation.
Degradome sequencing is a complex process which includes multiple steps such as isolating RNA fragments in a given sample as well as ligation
Ligation may refer to:
* Ligation (molecular biology), the covalent linking of two ends of DNA or RNA molecules
* In medicine, the making of a ligature (tie)
* Chemical ligation, the production of peptides from amino acids
* Tubal ligation, a meth ...
and reverse transcription to form complementary DNA (cDNA
In genetics, complementary DNA (cDNA) is DNA synthesized from a single-stranded RNA (e.g., messenger RNA (mRNA) or microRNA (miRNA)) template in a reaction catalyzed by the enzyme reverse transcriptase. cDNA is often used to express a speci ...
) strands. This cDNA can be sequenced, and the results are compared with a transcriptome
The transcriptome is the set of all RNA transcripts, including coding and non-coding, in an individual or a population of cells. The term can also sometimes be used to refer to all RNAs, or just mRNA, depending on the particular experiment. The t ...
, or reference genome, in order to determine and characterize the abundance of the RNA fragments identified in this process.
Methods

Technique
In general, the basic steps necessary for degradome sequencing include: # RNA isolation and separation: The RNA is isolated from the sample and size-fractionated to separate and classify small RNA molecules. # Library sequencing: The RNA is then ligated and used to form cDNA strands viareverse transcription
A reverse transcriptase (RT) is an enzyme used to generate complementary DNA (cDNA) from an RNA template, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, ...
. The cDNA is then denatured and replicated via polymerase chain reaction ( PCR). This yields a library generated and sequenced using high-throughput, deep sequencing methods.
# Analysis of data: The sequences yielded from these procedures are processed using bioinformatics tools to remove low-quality results, sequences that are adapters, and other forms of non-viable results. Following this screening step, the reads that remain are then compared with the reference genome (transcriptome) to determine the cleavage sites and their specific location.
# Analysis of cleavage sites: The cleavage sites are analyzed to identify the target transcripts of the enzyme that degrades RNA. Cleavage sites are identified in the 3' untranslated region ( UTR) of mRNAs. In addition, sequence motif
In biology, a sequence motif is a nucleotide or amino-acid sequence pattern that is widespread and usually assumed to be related to biological function of the macromolecule. For example, an ''N''-glycosylation site motif can be defined as ''As ...
s and other targets involved in miRNA-mediated degradation can be identified via the analysis of cleavage sites.
Analysis of Sequenced Raw Data
When analyzing the raw data derived from degradome sequencing, software tools like CleaveLand, PAREsnip, and miRferno are beneficial resources for researchers.CleaveLand Data Analysis Methodology
Degradome sequencing data and structural RNAs are used to remove all degradome sequences with exact matches to structural RNAs. The cDNA database is then used to map degradome sequences to cDNA sequences. The degradome sequences with many transcriptome hits are normalized. Then, query sequences of mRNA are generated for the matching degradome sequence. These query sequences are mapped tosmall RNA
Small RNA (sRNA) are polymeric RNA molecules that are less than 200 nucleotides in length, and are usually non-coding
Non-coding DNA (ncDNA) sequences are components of an organism's DNA that do not encode protein sequences. Some non-coding DNA ...
s, and a complementarity search is performed to match query sequences to small RNAs. A signal is then released to initiate noise analysis which works to distinguish and separate spurious results from real targets. Lastly, the resulting output of data analysis includes a list of all mRNA targets with the associated alignments for the small RNA-mRNA pairs.
Applications
Introduction
 The applications of degradome sequencing include identifying
The applications of degradome sequencing include identifying microRNA
MicroRNA (miRNA) are small, single-stranded, non-coding RNA molecules containing 21 to 23 nucleotides. Found in plants, animals and some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. miRN ...
(miRNA) targets, establishing mRNA methods of decay, and finding novel non-coding RNA fragments. In particular, this tool has been used to determine miRNA targets in numerous organisms, such as plants and mammals. Degradome sequencing has also been used to study the role of RNA decay pathways in cancer and identify new types of non-coding RNAs.
Ultimately, degradome sequencing is a powerful tool for the comprehensive analysis of RNA degradation with a variety of applications in biological research as well as medicine.
MicroRNA Research
MicroRNAs are a class of smallnoncoding RNA
A non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non- ...
created by removing stem-loop
Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when ...
precursors. MiRNAs play a role in controlling gene expression post-transcriptionally in addition to during transcription via RNA silencing. In order to accomplish this, the RNA-induced silencing complex (RISC
In computer engineering, a reduced instruction set computer (RISC) is a computer designed to simplify the individual instructions given to the computer to accomplish tasks. Compared to the instructions given to a complex instruction set comput ...
) processes pre-microRNAs into mature microRNAs. Mature miRNAs target specific mRNA species for regulation, often via the RISC complex disassembling specific mRNA sequences to inhibit translation
Translation is the communication of the Meaning (linguistic), meaning of a #Source and target languages, source-language text by means of an Dynamic and formal equivalence, equivalent #Source and target languages, target-language text. The ...
.
MiRNAs are highly conserved across a variety of species, so degradome sequencing is used in research to identify mRNA targets in many species. Degradome sequencing has been used to identify miRNA cleavage sites, because miRNAs can cause endonucleolytic cleavage of mRNA by extensive and often perfect complementarity to mRNAs. Degradome sequencing revealed many known and novel plant miRNA and small interfering RNA (siRNA
Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of double-stranded RNA at first non-coding RNA molecules, typically 20-24 (normally 21) base pairs in length, similar to miRNA, and operating wi ...
) targets. Recently, degradome sequencing also has been applied to identify animal (human and mouse) miRNA-derived cleavages.
Tracking microRNA Processing Signals by Degradome Sequencing Data Analysis
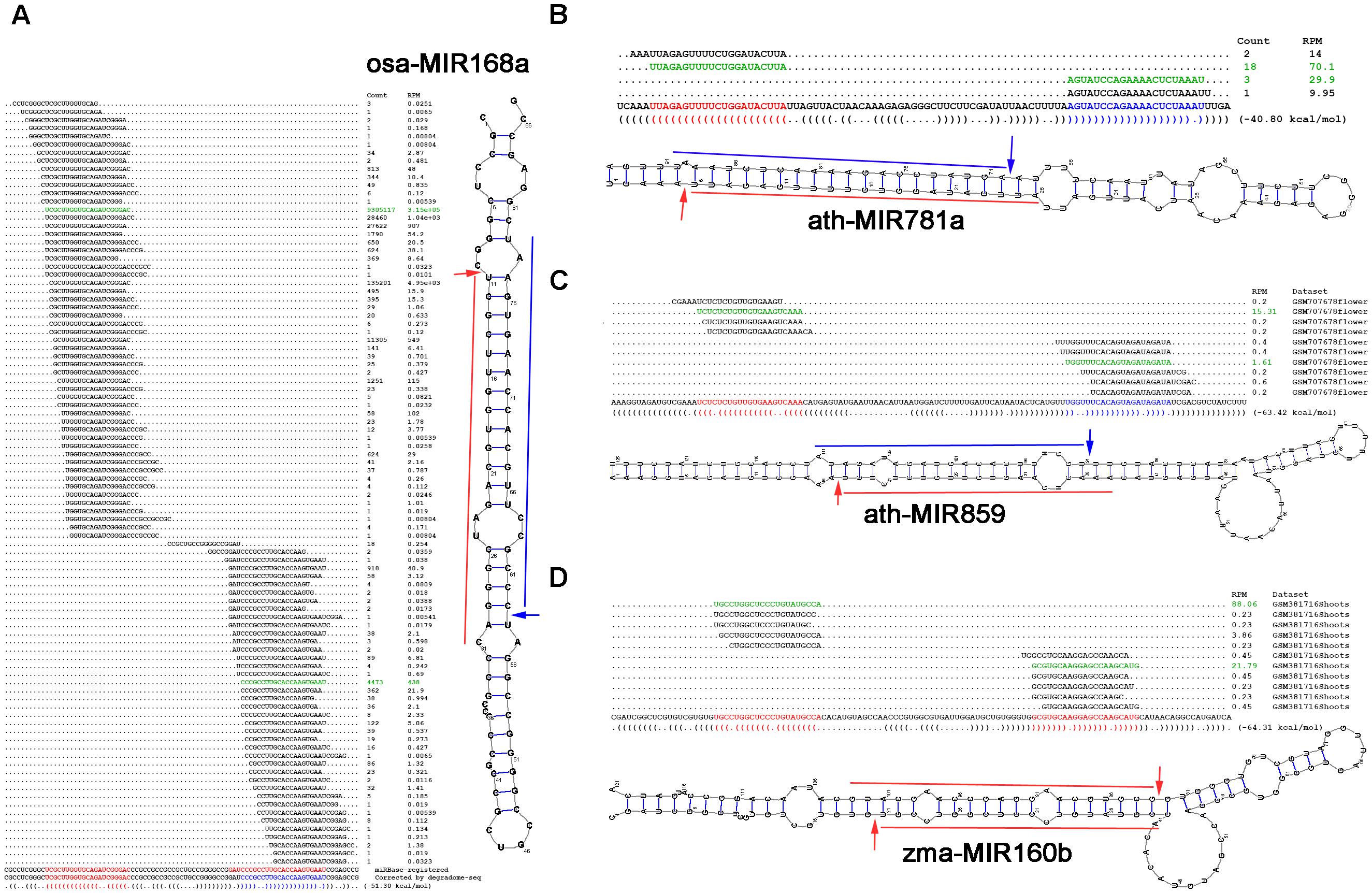
 In this study, researchers tracked and reported miRNA processing intermediates. Degradome signals on miRNA precursors were extracted and processed for 15 different species. The use of degradome sequencing in this study allowed for the collection of data that supported the analysis and processing of many miRNA precursors, with a greater ratio of high-confidence miRNAs annotated in
In this study, researchers tracked and reported miRNA processing intermediates. Degradome signals on miRNA precursors were extracted and processed for 15 different species. The use of degradome sequencing in this study allowed for the collection of data that supported the analysis and processing of many miRNA precursors, with a greater ratio of high-confidence miRNAs annotated in miRBase
In bioinformatics, miRBase is a biological database that acts as an archive of microRNA sequences and annotations. As of September 2010 it contained information about 15,172 microRNAs. This number has risen to 38,589 by March 2018. The miRBase re ...
, an miRNA database, than those considered low-confidence. Additionally, this study highlighted the importance of degradome sequencing as a technique in the study of miRNA annotation. In particular, the processing signal distribution provided by degradome sequencing data allowed the researchers to propose a new model for the method by which miRNAs are diced and to determine the frequency with which the loop-to-base mode of processing occurred. Ultimately, the results of this study are indicative of the impressive capability of degradome sequencing data to track miRNA processing signals, providing novel insights into miRNA processing and function.
The RNA Degradome: A Precious Resource for Deciphering RNA Processing and Regulation Codes in Plants
In this study, researchers developed a model in which biologists could use data derived from degradome sequencing to determine the effect of transcriptional and/or post-transcriptional regulation on patterns of gene expression in plants. In particular, this model applies degradome sequencing data to establish the method by whichsmall RNA
Small RNA (sRNA) are polymeric RNA molecules that are less than 200 nucleotides in length, and are usually non-coding
Non-coding DNA (ncDNA) sequences are components of an organism's DNA that do not encode protein sequences. Some non-coding DNA ...
s (sRNAs) mature and guide the process of targeted gene regulation. The results of this study demonstrate the vast potential applications of degradome sequencing analysis in future research regarding RNA biology in eukaryote
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacte ...
s. In particular, degradome sequencing data can be used to track non-coding RNA (ncRNA
A non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non-c ...
) processing signals which would be a valuable tool if expanded to include animal-based research.
External links
starBase database
a database for exploring microRNA cleavage sites from degradome sequencing (Degradome-Seq) data.
Cancer Research
Degradome sequencing can be used to identify cleavage sites of RNAs by sequencing the5' end
Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. In a single strand of DNA or RNA, the chemical convention of naming carbon atoms in the nucleotide pentose-sugar-ri ...
of the cleaved RNA fragments. This technique has been widely used in cancer
Cancer is a group of diseases involving abnormal cell growth with the potential to invade or spread to other parts of the body. These contrast with benign tumors, which do not spread. Possible signs and symptoms include a lump, abnormal b ...
research to identify potential targets of RNA-degrading enzymes involved in cancer progression. As such, degradome sequencing has provided a new method of discovering markers for earlier diagnosis and prognosis determination in cancer patients. Given the established role of extracellular protease
A protease (also called a peptidase, proteinase, or proteolytic enzyme) is an enzyme that catalyzes (increases reaction rate or "speeds up") proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the ...
s in promoting tumor development and growth across different tissues, degradome sequencing also holds important implications for discovering novel therapeutic targets for cancer treatments.

Degradome-Focused RNA Interference Screens to Identify Proteases Important for Breast Cancer Cell Growth
In this study, researchers utilized degradome sequencing to analyze all genome-encoded proteases involved in cell growth associated with breast cancer. Thesegenetic screen
A genetic screen or mutagenesis screen is an experimental technique used to identify and select individuals who possess a phenotype of interest in a mutagenized population. Hence a genetic screen is a type of phenotypic screen. Genetic screens c ...
s were performed in two breast cancer cell lines in mice which were phenotypically
In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology or physical form and structure, its developmental processes, its biochemical and physiological proper ...
distinct. One of these was a stem-cell like breast cancer cell line that altered its behavior under varied environmental conditions, such as the availability of oxygen and nutrients. Degradome sequencing, followed by a multistep selection process, revealed 100 protease genes that played a role in the growth of breast cancer cells. While the role of many of these protease genes in breast cancer growth was supported by previous research, this study found some proteases previously unknown to be involved in cancer growth. Additionally, this study revealed that environmental factors, such as nutrient and oxygen abundance, affect the extent to which breast cancer cells rely on specific proteases identified via degradome sequencing.
The results of this study were validated by using individual knockdown constructs in mice which functionally diminished the proteases of interest and affected the expression of breast cancer cells. These results indicate the high degree of reliability of degradome sequencing in identifying proteases involved in the growth of breast cancer cell lines in mouse models. Ultimately, this study concluded that degradome sequencing is a beneficial research tool for discovering and analyzing the functions of proteases in the proliferation of breast cancer. This holds many important implications for the potential degradome sequencing possesses as a diagnostic tool in early breast cancer detection and treatment.
References