|
Computational Genomics
Computational genomics refers to the use of computational and statistical analysis to decipher biology from genome sequences and related data, including both DNA and RNA sequence as well as other "post-genomic" data (i.e., experimental data obtained with technologies that require the genome sequence, such as genomic DNA microarrays). These, in combination with computational and statistical approaches to understanding the function of the genes and statistical association analysis, this field is also often referred to as Computational and Statistical Genetics/genomics. As such, computational genomics may be regarded as a subset of bioinformatics and computational biology, but with a focus on using whole genomes (rather than individual genes) to understand the principles of how the DNA of a species controls its biology at the molecular level and beyond. With the current abundance of massive biological datasets, computational studies have become one of the most important means to biolog ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome Sequence
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences (see non-coding DNA), and often a substantial fraction of 'junk' DNA with no evident function. Almost all eukaryotes have mitochondria and a small mitochondrial genome. Algae and plants also contain chloroplasts with a chloroplast genome. The study of the genome is called genomics. The genomes of many organisms have been sequenced and various regions have been annotated. The International Human Genome Project reported the sequence of the genome for ''Homo sapiens'' in 200The Human Genome Project although the initial "finished" sequence was missing 8% of the genome consisting mostly of repetitive sequences. With advancements in technology that could handle sequenc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intelligent Systems For Molecular Biology
Intelligent Systems for Molecular Biology (ISMB) is an annual academic conference on the subjects of bioinformatics and computational biology organised by the International Society for Computational Biology (ISCB). The principal focus of the conference is on the development and application of advanced computational methods for biological problems. The conference has been held every year since 1993 and has grown to become one of the largest and most prestigious meetings in these fields, hosting over 2,000 delegates in 2004. From the first meeting, ISMB has been held in locations worldwide; since 2007, meetings have been located in Europe and North America in alternating years. Since 2004, European meetings have been held jointly with the European Conference on Computational Biology (ECCB). The main ISMB conference is usually held over three days and consists of presentations, poster sessions and keynote talks. Most presentations are given in multiple parallel tracks; however, ke ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Minhash
In computer science and data mining, MinHash (or the min-wise independent permutations locality sensitive hashing scheme) is a technique for quickly estimating how similar two sets are. The scheme was invented by , and initially used in the AltaVista search engine to detect duplicate web pages and eliminate them from search results.. It has also been applied in large-scale clustering problems, such as clustering documents by the similarity of their sets of words.. Jaccard similarity and minimum hash values The Jaccard similarity coefficient is a commonly used indicator of the similarity between two sets. Let be a set and and be subsets of , then the Jaccard index is defined to be the ratio of the number of elements of their intersection and the number of elements of their union: : J(A,B) = . This value is 0 when the two sets are disjoint, 1 when they are equal, and strictly between 0 and 1 otherwise. Two sets are more similar (i.e. have relatively more members in common) w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Average Nucleotide Identity
Bacterial genomes are generally smaller and less variant in size among species when compared with genomes of eukaryotes. Bacterial genomes can range in size anywhere from about 130 kbp to over 14 Mbp. A study that included, but was not limited to, 478 bacterial genomes, concluded that as genome size increases, the number of genes increases at a disproportionately slower rate in eukaryotes than in non-eukaryotes. Thus, the proportion of non-coding DNA goes up with genome size more quickly in non-bacteria than in bacteria. This is consistent with the fact that most eukaryotic nuclear DNA is non-gene coding, while the majority of prokaryotic, viral, and organellar genes are coding. Right now, we have genome sequences from 50 different bacterial phyla and 11 different archaeal phyla. Second-generation sequencing has yielded many draft genomes (close to 90% of bacterial genomes in GenBank are currently not complete); third-generation sequencing might eventually yield a complete genome i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are typically represented as rows within a matrix. Gaps are inserted between the residues so that identical or similar characters are aligned in successive columns. Sequence alignments are also used for non-biological sequences, such as calculating the distance cost between strings in a natural language or in financial data. Interpretation If two sequences in an alignment share a common ancestor, mismatches can be interpreted as point mutations and gaps as indels (that is, insertion or deletion mutations) introduced in one or both lineages in the time since they diverged from one another. In sequence alignments of proteins, the degree of similarity between amino acids occupying a pa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Embryonic Development
An embryo is an initial stage of development of a multicellular organism. In organisms that reproduce sexually, embryonic development is the part of the life cycle that begins just after fertilization of the female egg cell by the male sperm cell. The resulting fusion of these two cells produces a single-celled zygote that undergoes many cell divisions that produce cells known as blastomeres. The blastomeres are arranged as a solid ball that when reaching a certain size, called a morula, takes in fluid to create a cavity called a blastocoel. The structure is then termed a blastula, or a blastocyst in mammals. The mammalian blastocyst hatches before implantating into the endometrial lining of the womb. Once implanted the embryo will continue its development through the next stages of gastrulation, neurulation, and organogenesis. Gastrulation is the formation of the three germ layers that will form all of the different parts of the body. Neurulation forms the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Conservation (genetics)
In evolutionary biology, conserved sequences are identical or similar sequences in nucleic acids ( DNA and RNA) or proteins across species ( orthologous sequences), or within a genome ( paralogous sequences), or between donor and receptor taxa ( xenologous sequences). Conservation indicates that a sequence has been maintained by natural selection. A highly conserved sequence is one that has remained relatively unchanged far back up the phylogenetic tree, and hence far back in geological time. Examples of highly conserved sequences include the RNA components of ribosomes present in all domains of life, the homeobox sequences widespread amongst Eukaryotes, and the tmRNA in Bacteria. The study of sequence conservation overlaps with the fields of genomics, proteomics, evolutionary biology, phylogenetics, bioinformatics and mathematics. History The discovery of the role of DNA in heredity, and observations by Frederick Sanger of variation between animal insulins in 1949, prompted ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Species
In biology, a species is the basic unit of Taxonomy (biology), classification and a taxonomic rank of an organism, as well as a unit of biodiversity. A species is often defined as the largest group of organisms in which any two individuals of the appropriate sexes or mating types can reproduction, produce Fertility, fertile offspring, typically by sexual reproduction. Other ways of defining species include their karyotype, DNA sequence, morphology (biology), morphology, behaviour or ecological niche. In addition, paleontologists use the concept of the chronospecies since fossil reproduction cannot be examined. The most recent rigorous estimate for the total number of species of eukaryotes is between 8 and 8.7 million. However, only about 14% of these had been described by 2011. All species (except viruses) are given a binomial nomenclature, two-part name, a "binomial". The first part of a binomial is the genus to which the species belongs. The second part is called the specifi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Comparative Genomics
Comparative genomics is a field of biological research in which the genomic features of different organisms are compared. The genomic features may include the DNA sequence, genes, gene order, regulatory sequences, and other genomic structural landmarks. In this branch of genomics, whole or large parts of genomes resulting from genome projects are compared to study basic biological similarities and differences as well as evolutionary relationships between organisms. The major principle of comparative genomics is that common features of two organisms will often be encoded within the DNA that is evolutionarily conserved between them. Therefore, comparative genomic approaches start with making some form of alignment of genome sequences and looking for orthologous sequences (sequences that share a common ancestry) in the aligned genomes and checking to what extent those sequences are conserved. Based on these, genome and molecular evolution are inferred and this may in turn be ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genes
In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and noncoding genes. During gene expression, the DNA is first copied into RNA. The RNA can be directly functional or be the intermediate template for a protein that performs a function. The transmission of genes to an organism's offspring is the basis of the inheritance of phenotypic traits. These genes make up different DNA sequences called genotypes. Genotypes along with environmental and developmental factors determine what the phenotypes will be. Most biological traits are under the influence of polygenes (many different genes) as well as g ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
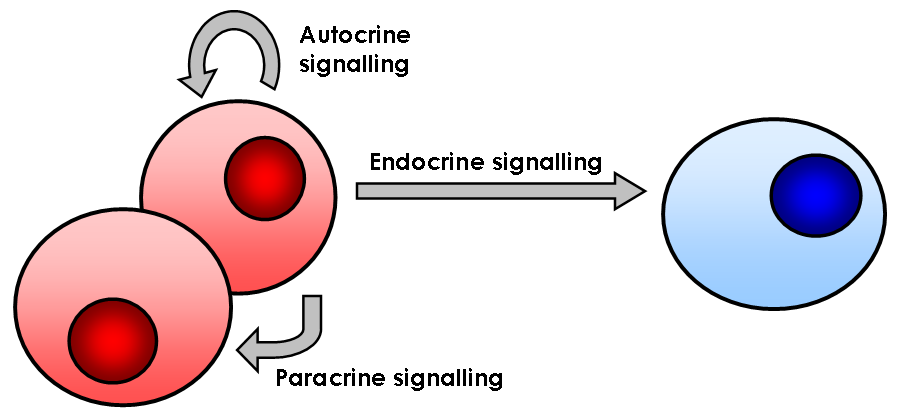
Cellular Signalling
In biology, cell signaling (cell signalling in British English) or cell communication is the ability of a cell to receive, process, and transmit signals with its environment and with itself. Cell signaling is a fundamental property of all cellular life in prokaryotes and eukaryotes. Signals that originate from outside a cell (or extracellular signals) can be physical agents like mechanical pressure, voltage, temperature, light, or chemical signals (e.g., small molecules, peptides, or gas). Cell signaling can occur over short or long distances, and as a result can be classified as autocrine, juxtacrine, intracrine, paracrine, or endocrine. Signaling molecules can be synthesized from various biosynthetic pathways and released through passive or active transports, or even from cell damage. Receptors play a key role in cell signaling as they are able to detect chemical signals or physical stimuli. Receptors are generally proteins located on the cell surface or within the interior ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication. The process of gene expression is used by all known life— eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea), and utilized by viruses—to generate the macromolecular machinery for life. In genetics, gene expression is the most fundamental level at which the genotype gives rise to the phen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |