|
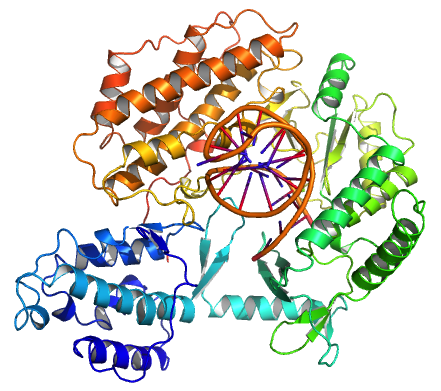
CST Complex
The CST complex is a cellular multiprotein complex involved in telomere maintenance. In budding yeast (''Saccharomyces cerevisiae''), it is composed of the proteins Cdc13, Stn1, and Ten1; in mammals, it consists of the proteins CTC1, STN1, and TEN1. It is related to the Replication protein A complex. Structure For budding yeast as well as for mammals, CST is a protein heterotrimer, consisting of three distinct proteins. Yeast Stn1 and Ten1 are orthologous proteins to mammalian STN1 and TEN1. But yeast Cdc13 and mammalian CTC1 are very different in amino acid sequence, length, and to some extent in function. Function For both budding yeast and mammals, the CST complex contributes to telomere maintenance, but this function is more crucial for budding yeast, where the CST complex performs the functions that shelterin performs in vertebrates. At least four factors contribute to telomere maintenance: telomerase, shelterin, TERRA and the CST Complex. CST protection of telomeres for ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Multiprotein Complex
A protein complex or multiprotein complex is a group of two or more associated polypeptide chains. Protein complexes are distinct from multienzyme complexes, in which multiple catalytic domains are found in a single polypeptide chain. Protein complexes are a form of quaternary structure. Proteins in a protein complex are linked by non-covalent protein–protein interactions. These complexes are a cornerstone of many (if not most) biological processes. The cell is seen to be composed of modular supramolecular complexes, each of which performs an independent, discrete biological function. Through proximity, the speed and selectivity of binding interactions between enzymatic complex and substrates can be vastly improved, leading to higher cellular efficiency. Many of the techniques used to enter cells and isolate proteins are inherently disruptive to such large complexes, complicating the task of determining the components of a complex. Examples of protein complexes include the p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TERRA (biology)
TERRA in biology is an abbreviation for "TElomeric Repeat-containing RNA". TERRA is RNA that is transcribed from telomeres — the repeating 6-nucleotide sequences that cap the ends of chromosomes. TERRA functions with shelterin to inhibit telomere lengthening by enzyme telomerase. TERRAs are essential for telomere length and maintenance. At least four factors contribute to telomere maintenance: telomerase, shelterin, TERRA and the CST Complex. TERRA can also regulate telomere length by increasing euchromatin formation. On the other hand, nonsense-mediated decay factor enrichment at telomeres may exist to prevent TERRA inhibition of telomerase. TERRA levels vary during the cell cycle, decreasing during S phase, and increasing in the transition from G2 phase to G1 phase The G1 phase, gap 1 phase, or growth 1 phase, is the first of four phases of the cell cycle that takes place in eukaryotic cell division. In this part of interphase, the cell synthesizes mRNA and proteins in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Shelterin
Shelterin (also called telosome) is a protein complex known to protect telomeres in many eukaryotes from DNA repair mechanisms, as well as to regulate telomerase activity. In mammals and other vertebrates, telomeric DNA consists of repeating double-stranded 5'-TTAGGG-3' (G-strand) sequences (2-15 kilobases in humans) along with the 3'-AATCCC-5' (C-strand) complement, ending with a 50-400 nucleotide 3' (G-strand) overhang. Much of the final double-stranded portion of the telomere forms a T-loop (Telomere-loop) that is invaded by the 3' (G-strand) overhang to form a small D-loop (Displacement-loop). The absence of shelterin causes telomere uncapping and thereby activates damage-signaling pathways that may lead to non-homologous end joining (NHEJ), homology directed repair (HDR), end-to-end fusions, genomic instability, senescence, or apoptosis. Subunits Shelterin has six subunits: TRF1, TRF2, POT1, RAP1, TIN2, and TPP1. They can operate in smaller subsets to re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Guanine
Guanine () ( symbol G or Gua) is one of the four main nucleobases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine (uracil in RNA). In DNA, guanine is paired with cytosine. The guanine nucleoside is called guanosine. With the formula C5H5N5O, guanine is a derivative of purine, consisting of a fused pyrimidine-imidazole ring system with conjugated double bonds. This unsaturated arrangement means the bicyclic molecule is planar. Properties Guanine, along with adenine and cytosine, is present in both DNA and RNA, whereas thymine is usually seen only in DNA, and uracil only in RNA. Guanine has two tautomeric forms, the major keto form (see figures) and rare enol form. It binds to cytosine through three hydrogen bonds. In cytosine, the amino group acts as the hydrogen bond donor and the C-2 carbonyl and the N-3 amine as the hydrogen-bond acceptors. Guanine has the C-6 carbonyl group that acts as the hydrogen bond acceptor, while a group at N ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleic Acid Tertiary Structure
Nucleic acid tertiary structure is the three-dimensional shape of a nucleic acid polymer. RNA and DNA molecules are capable of diverse functions ranging from molecular recognition to catalysis. Such functions require a precise three-dimensional structure. While such structures are diverse and seemingly complex, they are composed of recurring, easily recognizable tertiary structural motifs that serve as molecular building blocks. Some of the most common motifs for RNA and DNA tertiary structure are described below, but this information is based on a limited number of solved structures. Many more tertiary structural motifs will be revealed as new RNA and DNA molecules are structurally characterized. Helical structures Double helix The double helix is the dominant tertiary structure for biological DNA, and is also a possible structure for RNA. Three DNA conformations are believed to be found in nature, A-DNA, B-DNA, and Z-DNA. The "B" form described by James D. Wats ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
G-quadruplex
In molecular biology, G-quadruplex secondary structures (G4) are formed in nucleic acids by sequences that are rich in guanine. They are helical in shape and contain guanine tetrads that can form from one, two or four strands. The unimolecular forms often occur naturally near the ends of the chromosomes, better known as the telomeric regions, and in transcriptional regulatory regions of multiple genes, both in microbes and across vertebrates including oncogenes in humans. Four guanine bases can associate through Hoogsteen hydrogen bonding to form a square planar structure called a guanine tetrad (G-tetrad or G-quartet), and two or more guanine tetrads (from G-tracts, continuous runs of guanine) can stack on top of each other to form a G-quadruplex. The placement and bonding to form G-quadruplexes is not random and serve very unusual functional purposes. The quadruplex structure is further stabilized by the presence of a cation, especially potassium, which sits in a central c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
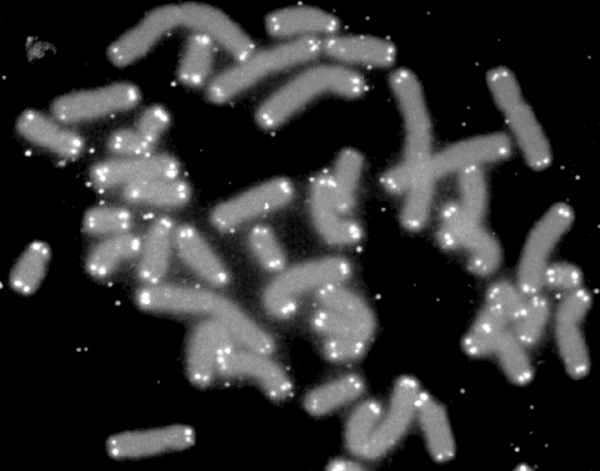
Telomere
A telomere (; ) is a region of repetitive nucleotide sequences associated with specialized proteins at the ends of linear chromosomes. Although there are different architectures, telomeres, in a broad sense, are a widespread genetic feature most commonly found in eukaryotes. In most, if not all species possessing them, they protect the terminal regions of chromosomal DNA from progressive degradation and ensure the integrity of linear chromosomes by preventing DNA repair systems from mistaking the very ends of the DNA strand for a double-strand break. Discovery In the early 1970s, Soviet theorist Alexei Olovnikov first recognized that chromosomes could not completely replicate their ends; this is known as the "end replication problem". Building on this, and accommodating Leonard Hayflick's idea of limited somatic cell division, Olovnikov suggested that DNA sequences are lost every time a cell replicates until the loss reaches a critical level, at which point cell division end ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritance. This is essential for cell division during growth and repair of damaged tissues, while it also ensures that each of the new cells receives its own copy of the DNA. The cell possesses the distinctive property of division, which makes replication of DNA essential. DNA is made up of a double helix of two complementary strands. The double helix describes the appearance of a double-stranded DNA which is thus composed of two linear strands that run opposite to each other and twist together to form. During replication, these strands are separated. Each strand of the original DNA molecule then serves as a template for the production of its counterpart, a process referred to as semiconservative replication. As a result of semi-conservative rep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Replication Stress
DNA replication stress refers to the state of a cell whose genome is exposed to various stresses. The events that contribute to replication stress occur during DNA replication, and can result in a stalled replication fork. There are many events that contribute to replication stress, including: * Misincorporation of ribonucleotides * Unusual DNA structures * Conflicts between replication and transcription * Insufficiency of essential replication factors * Common fragile sites * Overexpression or constitutive activation of oncogenes * Chromatin inaccessibility ATM and ATR are proteins that help to alleviate replication stress. Specifically, they are kinases that are recruited and activated by DNA damage. The stalled replication fork can collapse if these regulatory proteins fail to stabilize it. When this occurs, reassembly of the fork is initiated in order to repair the damaged DNA end. Replication fork The replication fork consists of a group of proteins that influence th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Telomerase
Telomerase, also called terminal transferase, is a ribonucleoprotein that adds a species-dependent telomere repeat sequence to the 3' end of telomeres. A telomere is a region of repetitive sequences at each end of the chromosomes of most eukaryotes. Telomeres protect the end of the chromosome from DNA damage or from fusion with neighbouring chromosomes. The fruit fly ''Drosophila melanogaster'' lacks telomerase, but instead uses retrotransposons to maintain telomeres. Telomerase is a reverse transcriptase enzyme that carries its own RNA molecule (e.g., with the sequence 3′- CCC AA UCCC-5′ in ''Trypanosoma brucei'') which is used as a template when it elongates telomeres. Telomerase is active in gametes and most cancer cells, but is normally absent from, or at very low levels in, most somatic cells. History The existence of a compensatory mechanism for telomere shortening was first found by Soviet biologist Alexey Olovnikov in 1973, who also suggested the telomere hypothe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Telomere
A telomere (; ) is a region of repetitive nucleotide sequences associated with specialized proteins at the ends of linear chromosomes. Although there are different architectures, telomeres, in a broad sense, are a widespread genetic feature most commonly found in eukaryotes. In most, if not all species possessing them, they protect the terminal regions of chromosomal DNA from progressive degradation and ensure the integrity of linear chromosomes by preventing DNA repair systems from mistaking the very ends of the DNA strand for a double-strand break. Discovery In the early 1970s, Soviet theorist Alexei Olovnikov first recognized that chromosomes could not completely replicate their ends; this is known as the "end replication problem". Building on this, and accommodating Leonard Hayflick's idea of limited somatic cell division, Olovnikov suggested that DNA sequences are lost every time a cell replicates until the loss reaches a critical level, at which point cell division end ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Shelterin
Shelterin (also called telosome) is a protein complex known to protect telomeres in many eukaryotes from DNA repair mechanisms, as well as to regulate telomerase activity. In mammals and other vertebrates, telomeric DNA consists of repeating double-stranded 5'-TTAGGG-3' (G-strand) sequences (2-15 kilobases in humans) along with the 3'-AATCCC-5' (C-strand) complement, ending with a 50-400 nucleotide 3' (G-strand) overhang. Much of the final double-stranded portion of the telomere forms a T-loop (Telomere-loop) that is invaded by the 3' (G-strand) overhang to form a small D-loop (Displacement-loop). The absence of shelterin causes telomere uncapping and thereby activates damage-signaling pathways that may lead to non-homologous end joining (NHEJ), homology directed repair (HDR), end-to-end fusions, genomic instability, senescence, or apoptosis. Subunits Shelterin has six subunits: TRF1, TRF2, POT1, RAP1, TIN2, and TPP1. They can operate in smaller subsets to re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |