|
COVID-19 Testing
COVID-19 testing involves analyzing samples to assess the current or past presence of SARS-CoV-2, the virus that cases COVID-19 and is responsible for the COVID-19 pandemic. The two main types of tests detect either the presence of the virus or antibodies produced in response to infection. Molecular tests for viral presence through its molecular components are used to diagnose individual cases and to allow public health authorities to trace and contain outbreaks. Antibody tests (serology immunoassays) instead show whether someone once had the disease. They are less useful for diagnosing current infections because antibodies may not develop for weeks after infection. It is used to assess disease prevalence, which aids the estimation of the infection fatality rate. Individual jurisdictions have adopted varied testing protocols, including whom to test, how often to test, analysis protocols, sample collection and the uses of test results. This variation has likely significantly ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
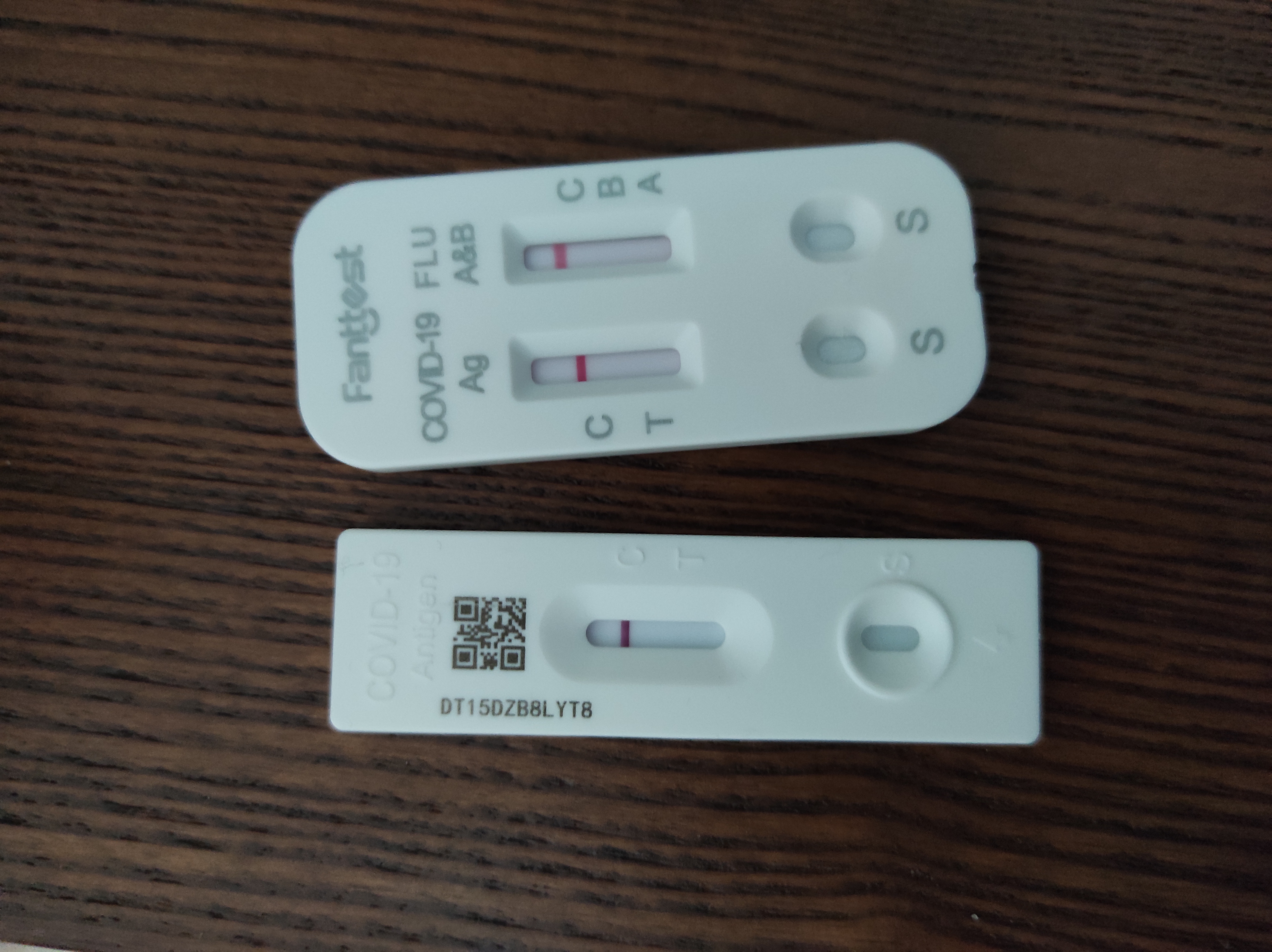
Rapid Antigen Test
A rapid antigen test (RAT), sometimes called a rapid antigen detection test (RADT), antigen rapid test (ART), or wikt:Appendix:Glossary#loosely, loosely just a rapid test, is a rapid diagnostic test suitable for point-of-care testing that directly detects the presence or absence of an antigen. RATs are a type of lateral flow test detecting antigens, rather than antibody, antibodies (Antibody titer, antibody tests) or nucleic acid (nucleic acid tests). Rapid tests generally give a result in 5 to 30 minutes, require minimal training or infrastructure, and have significant cost advantages. Rapid antigen tests for the detection of SARS-CoV-2, the virus that causes COVID-19, have been commonly used during the COVID-19 pandemic. For many years, an early and major class of RATs—the rapid strep tests for streptococci—were so often the referent when RATs or RADTs were mentioned that the two latter terms were often loosely treated as synonymous with those. Since the COVID-19 pandemic, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CDC 2019-nCoV Laboratory Test Kit
The Centers for Disease Control and Prevention (CDC) is the National public health institutes, national public health agency of the United States. It is a Federal agencies of the United States, United States federal agency under the United States Department of Health and Human Services, Department of Health and Human Services (HHS), and is headquartered in Atlanta, Georgia. The CDC's current nominee for director is Susan Monarez. She became acting director on January 23, 2025, but stepped down on March 24, 2025 when nominated for the director position. On May 14, 2025, Robert F. Kennedy Jr. stated that lawyer Matthew Buzzelli is acting CDC director. However, the CDC web site does not state the acting director's name. The agency's main goal is the protection of public health and safety through the control and prevention of disease, injury, and disability in the US and worldwide. The CDC focuses national attention on developing and applying disease control and prevention. It e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Helsinki
Helsinki () is the Capital city, capital and most populous List of cities and towns in Finland, city in Finland. It is on the shore of the Gulf of Finland and is the seat of southern Finland's Uusimaa region. About people live in the municipality, with million in the Helsinki capital region, capital region and million in the Helsinki metropolitan area, metropolitan area. As the most populous List of urban areas in Finland by population, urban area in Finland, it is the country's most significant centre for politics, education, finance, culture, and research. Helsinki is north of Tallinn, Estonia, east of Stockholm, Sweden, and west of Saint Petersburg, Russia. Helsinki has significant History of Helsinki, historical connections with these three cities. Together with the cities of Espoo, Vantaa and Kauniainen—and surrounding commuter towns, including the neighbouring municipality of Sipoo to the east—Helsinki forms a Helsinki metropolitan area, metropolitan are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Minimum Information For Publication Of Quantitative Real-Time PCR Experiments
The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines are a set of protocols for conducting and reporting quantitative real-time PCR experiments and data, as devised by Bustin et al. in 2009. They were devised after a paper was published in 2002 that claimed to detect measles virus in children with autism through the use of RT-qPCR, but the results proved to be completely unreproducible by other scientists. The authors themselves also did not try to reproduce them and the raw data was found to have a large amount of errors and basic mistakes in analysis. This incident prompted Stephen Bustin to create the MIQE guidelines to provide a baseline level of quality for qPCR data published in scientific literature. Purpose The MIQE guidelines were created due to the low quality of qPCR data submitted to academic journals at the time, which was only becoming more common as Next Generation Sequencing machinery allowed for such experiments to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
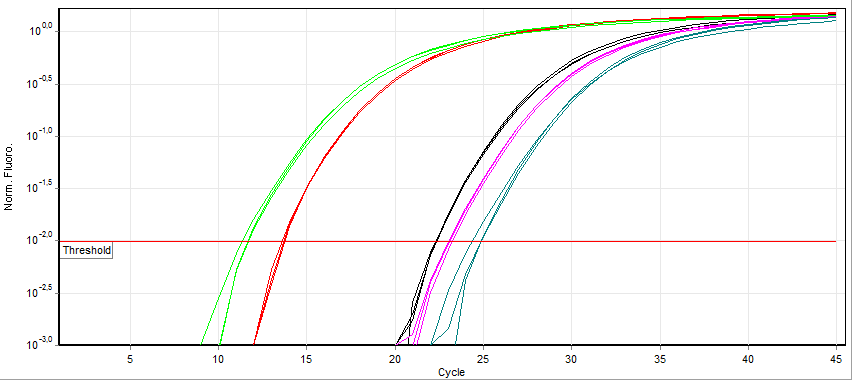
Real-time Polymerase Chain Reaction
A real-time polymerase chain reaction (real-time PCR, or qPCR when used quantitatively) is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR (i.e., in real time), not at its end, as in conventional PCR. Real-time PCR can be used quantitatively and semi-quantitatively (i.e., above/below a certain amount of DNA molecules). Two common methods for the detection of PCR products in real-time PCR are (1) non-specific fluorescent dyes that Intercalation (biochemistry), intercalate with any double-stranded DNA and (2) sequence-specific DNA probes consisting of oligonucleotides that are labelled with a fluorescence, fluorescent reporter, which permits detection only after nucleic acid hybridisation, hybridization of the probe with its complementary sequence. The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines propose that the abb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reverse Transcription Polymerase Chain Reaction
Reverse transcription polymerase chain reaction (RT-PCR) is a laboratory technique combining reverse transcription of RNA into DNA (in this context called complementary DNA or cDNA) and amplification of specific DNA targets using polymerase chain reaction (PCR). It is primarily used to measure the amount of a specific RNA. This is achieved by monitoring the amplification reaction using fluorescence, a technique called real-time PCR or quantitative PCR (qPCR). Confusion can arise because some authors use the acronym RT-PCR to denote real-time PCR. In this article, RT-PCR will denote Reverse Transcription PCR. Combined RT-PCR and qPCR are routinely used for analysis of gene expression and quantification of viral RNA in research and clinical settings. The close association between RT-PCR and qPCR has led to metonymic use of the term qPCR to mean RT-PCR. Such use may be confusing, as RT-PCR can be used without qPCR, for example to enable molecular cloning, sequencing or simple det ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reverse Transcriptase
A reverse transcriptase (RT) is an enzyme used to convert RNA genome to DNA, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobile genetic elements to proliferate within the host genome, and by eukaryotic cells to extend the telomeres at the ends of their linear chromosomes. The process does not violate the flows of genetic information as described by the classical central dogma, but rather expands it to include transfers of information from RNA to DNA. Retroviral RT has three sequential biochemical activities: RNA-dependent DNA polymerase activity, ribonuclease H (RNase H), and DNA-dependent DNA polymerase activity. Collectively, these activities enable the enzyme to convert single-stranded RNA into double-stranded cDNA. In retroviruses and retrotransposons, this cDNA can then integrate into the host genome, from which new RNA copies can be made via host-cell ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all life, living organisms, acting as the most essential part of heredity, biological inheritance. This is essential for cell division during growth and repair of damaged tissues, while it also ensures that each of the new cells receives its own copy of the DNA. The cell possesses the distinctive property of division, which makes replication of DNA essential. DNA is made up of a nucleic acid double helix, double helix of two Complementary DNA, complementary DNA strand, strands. DNA is often called double helix. The double helix describes the appearance of a double-stranded DNA which is composed of two linear strands that run opposite to each other and twist together. During replication, these strands are separated. Each strand of the original DNA molecule then serves as a template for the production of its counterpart, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA (or a part of it) sufficiently to enable detailed study. PCR was invented in 1983 by American biochemist Kary Mullis at Cetus Corporation. Mullis and biochemist Michael Smith (chemist), Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993. PCR is fundamental to many of the procedures used in genetic testing and research, including analysis of Ancient DNA, ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory research for a broad variety of applications including biomedical research and forensic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Massive Parallel Sequencing
Massive parallel sequencing or massively parallel sequencing is any of several high-throughput approaches to DNA sequencing using the concept of massively parallel processing; it is also called next-generation sequencing (NGS) or second-generation sequencing. Some of these technologies emerged between 1993 and 1998 and have been commercially available since 2005. These technologies use miniaturized and parallelized platforms for sequencing of 1 million to 43 billion short reads (50 to 400 bases each) per instrument run. Many NGS platforms differ in engineering configurations and sequencing chemistry. They share the technical paradigm of massive parallel sequencing via spatially separated, clonally amplified DNA templates or single DNA molecules in a flow cell. This design is very different from that of Sanger sequencing—also known as capillary sequencing or first-generation sequencing—which is based on electrophoretic separation of chain-termination products produced in ind ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microarray Analysis Techniques
Microarray analysis techniques are used in interpreting the data generated from experiments on DNA (Gene chip analysis), RNA, and protein microarrays, which allow researchers to investigate the expression state of a large number of genesin many cases, an organism's entire genomein a single experiment. Such experiments can generate very large amounts of data, allowing researchers to assess the overall state of a cell or organism. Data in such large quantities is difficultif not impossibleto analyze without the help of computer programs. Introduction Microarray data analysis is the final step in reading and processing data produced by a microarray chip. Samples undergo various processes including purification and scanning using the microchip, which then produces a large amount of data that requires processing via computer software. It involves several distinct steps, as outlined in the image below. Changing any one of the steps will change the outcome of the analysis, so the MAQC Proj ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Digital Polymerase Chain Reaction
Digital polymerase chain reaction (digital PCR, DigitalPCR, dPCR, or dePCR) is a biotechnological refinement of conventional polymerase chain reaction methods that can be used to directly quantify and clonally amplify nucleic acids strands including DNA, cDNA, or RNA. The key difference between dPCR and qPCR lies in the method of measuring nucleic acids amounts, with the former being a more precise method than PCR, though also more prone to error in the hands of inexperienced users. PCR carries out one reaction per single sample. dPCR also carries out a single reaction within a sample, however the sample is separated into a large number of partitions and the reaction is carried out in each partition individually. This separation allows a more reliable collection and sensitive measurement of nucleic acid amounts. The method has been demonstrated as useful for studying variations in gene sequences—such as copy number variants and point mutations. Principles The polymerase cha ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |