|
Barnase
Barnase (a portmanteau of "BActerial" "RiboNucleASE") is a bacterial protein that consists of 110 amino acids and has ribonuclease activity. It is synthesized and secreted by the bacterium ''Bacillus amyloliquefaciens'', but is lethal to the cell when expressed without its inhibitor barstar. The inhibitor binds to and occludes the ribonuclease active site, preventing barnase from damaging the cell's RNA after it has been synthesized but before it has been secreted. The barnase/barstar complex is noted for its extraordinarily tight protein-protein binding, with an on-rate of 108s−1M−1. Protein folding studies Barnase has no disulfide bonds, nor does it require divalent cations or non-peptide components to fold. This simplicity, in combination with its reversible folding transition, means that barnase has been extensively studied in order to understand how proteins fold. The folding of barnase has been extensively studied in the laboratory of Alan Fersht, who used it as the te ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Barstar
Barstar is a small protein synthesized by the bacterium ''Bacillus amyloliquefaciens''. Its function is to inhibit the ribonuclease activity of its binding partner barnase Barnase (a portmanteau of "BActerial" "RiboNucleASE") is a bacterial protein that consists of 110 amino acids and has ribonuclease activity. It is synthesized and secreted by the bacterium '' Bacillus amyloliquefaciens'', but is lethal to the cel ..., with which it forms an extraordinarily tightly bound complex within the cell until barnase is secreted. Expression of barstar is necessary to counter the lethal effect of expressed active barnase. The structure of the barnase-barstar complex is known. References Bacterial proteins {{protein-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alan Fersht
Sir Alan Roy Fersht (born 21 April 1943) is a British chemist at the MRC Laboratory of Molecular Biology, Cambridge, and an Emeritus Professor in the Department of Chemistry at the University of Cambridge. He was Master of Gonville and Caius College, Cambridge from 2012 to 2018. He works on protein folding, and is sometimes described as a founder of protein engineering. Early life and education Fersht was born on 21 April 1943 in Hackney, London. His father, Philip, was a ladies' tailor and his mother, Betty, a dressmaker. His grandparents were Jewish immigrants from Poland, Romania, Lithuania and Belarus. He was educated at Sir George Monoux Grammar School, an all-boys grammar school in Walthamstow, London. He was a keen chess player and was the Essex County Junior champion in 1961. He was awarded a State Scholarship to read Natural Sciences at Gonville and Caius College, Cambridge, where he obtained First Class in Pt I of the Natural Sciences Tripos in 1964, First Class i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Toxin-antitoxin System
A toxin-antitoxin system is a set of two or more closely linked genes that together encode both a "toxin" protein and a corresponding "antitoxin". Toxin-antitoxin systems are widely distributed in prokaryotes, and organisms often have them in multiple copies. When these systems are contained on plasmids – transferable genetic elements – they ensure that only the daughter cells that inherit the plasmid survive after cell division. If the plasmid is absent in a daughter cell, the unstable antitoxin is degraded and the stable toxic protein kills the new cell; this is known as 'post-segregational killing' (PSK). Toxin-antitoxin systems are typically classified according to how the antitoxin neutralises the toxin. In a type I toxin-antitoxin system, the translation of messenger RNA (mRNA) that encodes the toxin is inhibited by the binding of a small non-coding RNA antitoxin that binds the toxin mRNA. The toxic protein in a type II system is inhibited post-translationally ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
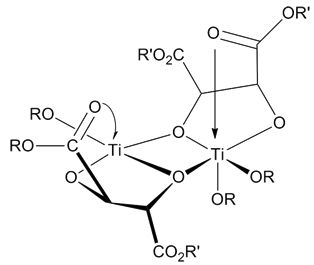
Transesterification
In organic chemistry, transesterification is the process of exchanging the organic group R″ of an ester with the organic group R' of an alcohol. These reactions are often catalyzed by the addition of an acid or base catalyst. The reaction can also be accomplished with the help of other enzymes, particularly lipases (one example is the lipase E.C.3.1.1.3). Strong acids catalyse the reaction by donating a proton to the carbonyl group, thus making it a more potent electrophile, whereas bases catalyse the reaction by removing a proton from the alcohol, thus making it more nucleophilic. If the alcohol produced by the reaction can be separated from the reactants by distillation this will drive the equilibrium toward the products, this means that esters with larger alkoxy groups can be made from methyl or ethyl esters in high purity by heating the mixture of ester, acid/base, and large alcohol. Mechanism In the transesterification mechanism, the carbonyl carbon of the starting e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme Catalysis
Enzyme catalysis is the increase in the rate of a process by a biological molecule, an "enzyme". Most enzymes are proteins, and most such processes are chemical reactions. Within the enzyme, generally catalysis occurs at a localized site, called the active site. Most enzymes are made predominantly of proteins, either a single protein chain or many such chains in a multi-subunit complex. Enzymes often also incorporate non-protein components, such as metal ions or specialized organic molecules known as cofactor (e.g. adenosine triphosphate). Many cofactors are vitamins, and their role as vitamins is directly linked to their use in the catalysis of biological process within metabolism. Catalysis of biochemical reactions in the cell is vital since many but not all metabolically essential reactions have very low rates when uncatalysed. One driver of protein evolution is the optimization of such catalytic activities, although only the most crucial enzymes operate near catalytic e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrolysis
Hydrolysis (; ) is any chemical reaction in which a molecule of water breaks one or more chemical bonds. The term is used broadly for substitution reaction, substitution, elimination reaction, elimination, and solvation reactions in which water is the nucleophile. Biological hydrolysis is the cleavage of biomolecules where a water molecule is consumed to effect the separation of a larger molecule into component parts. When a carbohydrate is broken into its component sugar molecules by hydrolysis (e.g., sucrose being broken down into glucose and fructose), this is recognized as saccharification. Hydrolysis reactions can be the reverse of a condensation reaction in which two molecules join into a larger one and eject a water molecule. Thus hydrolysis adds water to break down, whereas condensation builds up by removing water. Types Usually hydrolysis is a chemical process in which a molecule of water is added to a substance. Sometimes this addition causes both the substance and w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phi Value Analysis
Phi value analysis, \phi analysis, or \phi-value analysis is an experimental protein engineering technique for studying the structure of the folding transition state of small protein domains that fold in a two-state manner. The structure of the folding transition state is hard to find using methods such as protein NMR or X-ray crystallography because folding transitions states are mobile and partly unstructured by definition. In \phi -value analysis, the folding kinetics and conformational folding stability of the wild-type protein are compared with those of point mutants to find ''phi values''. These measure the mutant residue's energetic contribution to the folding transition state, which reveals the degree of native structure around the mutated residue in the transition state, by accounting for the relative free energies of the unfolded state, the folded state, and the transition state for the wild-type and mutant proteins. The protein's residues are mutated one by one to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transition State
In chemistry, the transition state of a chemical reaction is a particular configuration along the reaction coordinate. It is defined as the state corresponding to the highest potential energy along this reaction coordinate. It is often marked with the double dagger ‡ symbol. As an example, the transition state shown below occurs during the SN2 reaction of bromoethane with a hydroxide anion: The activated complex of a reaction can refer to either the transition state or to other states along the reaction coordinate between reactants and products, especially those close to the transition state.Peter Atkins and Julio de Paula, ''Physical Chemistry'' (8th ed., W.H. Freeman 2006), p.809 According to the transition state theory, once the reactants have passed through the transition state configuration, they always continue to form products. History of concept The concept of a transition state has been important in many theories of the rates at which chemical reactions occ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Folding
Protein folding is the physical process by which a protein chain is translated to its native three-dimensional structure, typically a "folded" conformation by which the protein becomes biologically functional. Via an expeditious and reproducible process, a polypeptide folds into its characteristic three-dimensional structure from a random coil. Each protein exists first as an unfolded polypeptide or random coil after being translated from a sequence of mRNA to a linear chain of amino acids. At this stage the polypeptide lacks any stable (long-lasting) three-dimensional structure (the left hand side of the first figure). As the polypeptide chain is being synthesized by a ribosome, the linear chain begins to fold into its three-dimensional structure. Folding of many proteins begins even during translation of the polypeptide chain. Amino acids interact with each other to produce a well-defined three-dimensional structure, the folded protein (the right hand side of the figure), ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Divalent Cations
In chemistry, the valence (US spelling) or valency (British spelling) of an element is the measure of its combining capacity with other atoms when it forms chemical compounds or molecules. Description The combining capacity, or affinity of an atom of a given element is determined by the number of hydrogen atoms that it combines with. In methane, carbon has a valence of 4; in ammonia, nitrogen has a valence of 3; in water, oxygen has a valence of 2; and in hydrogen chloride, chlorine has a valence of 1. Chlorine, as it has a valence of one, can be substituted for hydrogen. Phosphorus has a valence of 5 in phosphorus pentachloride, . Valence diagrams of a compound represent the connectivity of the elements, with lines drawn between two elements, sometimes called bonds, representing a saturated valency for each element. The two tables below show some examples of different compounds, their valence diagrams, and the valences for each element of the compound. Modern definitions ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the three dimensional conformational isomerism, form of ''local segments'' of proteins. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Disulfide Bonds
In biochemistry, a disulfide (or disulphide in British English) refers to a functional group with the structure . The linkage is also called an SS-bond or sometimes a disulfide bridge and is usually derived by the coupling of two thiol groups. In biology, disulfide bridges formed between thiol groups in two cysteine residues are an important component of the secondary and tertiary structure of protein, proteins. ''Persulfide'' usually refers to compounds. In inorganic chemistry disulfide usually refers to the corresponding anion (−S−S−). Organic disulfides Symmetrical disulfides are compounds of the formula . Most disulfides encountered in organo sulfur chemistry are symmetrical disulfides. Unsymmetrical disulfides (also called heterodisulfides) are compounds of the formula . They are less common in organic chemistry, but most disulfides in nature are unsymmetrical. Properties The disulfide bonds are strong, with a typical bond dissociation energy of 60 kcal/mol ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |