|
Arsenate Reductase (azurin)
Arsenate reductase (azurin) () is an enzyme that catalysis, catalyzes the chemical reaction :arsenite + H2O + 2 azurinox \rightleftharpoons arsenate + 2 azurinred + 2 H+ The 3 substrate (biochemistry), substrates of this enzyme are arsenite, water, and oxidised azurin, whereas its 3 product (chemistry), products are arsenate, reduced azurin, and hydrogen ion. Classification This enzyme belongs to the family of oxidoreductases, specifically those acting on phosphorus or arsenic in donor with a copper protein as acceptor. Nomenclature The List of enzymes, systematic name of this enzyme class is arsenite:azurin oxidoreductase. This enzyme is also called arsenite oxidase. Structure and function The enzyme contains a molybdopterin centre comprising two molybdopterin guanosine dinucleotide cofactors bound to molybdenum, a Iron-sulfur protein, [3Fe-4S] cluster and a Rieske protein, Rieske-type [2Fe-2S] cluster. Also uses a c-type cytochrome or as acceptors. References * * ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the react ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Copper Protein
Copper proteins are proteins that contain one or more copper ions as prosthetic groups. Copper proteins are found in all forms of air-breathing life. These proteins are usually associated with electron-transfer with or without the involvement of oxygen (O2). Some organisms even use copper proteins to carry oxygen instead of iron proteins. A prominent copper proteins in humans is in cytochrome c oxidase (cco). The enzyme cco mediates the controlled combustion that produces ATP. Classes The metal centers in the copper proteins can be classified into several types: *Type I copper centres (T1Cu) are characterized by a single copper atom coordinated by two histidine residues and a cysteine residue in a trigonal planar structure, and a variable axial ligand. In class I T1Cu proteins (e.g. amicyanin, plastocyanin and pseudoazurin) the axial ligand is the sulfur of methionine, whereas aminoacids other than methionine (e.g. glutamine) give rise to class II T1Cu copper protei ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cytochrome
Cytochromes are redox-active proteins containing a heme, with a central Fe atom at its core, as a cofactor. They are involved in electron transport chain and redox catalysis. They are classified according to the type of heme and its mode of binding. Four varieties are recognized by the International Union of Biochemistry and Molecular Biology (IUBMB), cytochromes a, cytochromes b, cytochromes c and cytochrome d. Cytochrome function is linked to the reversible redox change from ferrous (Fe(II)) to the ferric (Fe(III)) oxidation state of the iron found in the heme core. In addition to the classification by the IUBMB into four cytochrome classes, several additional classifications such as cytochrome o and cytochrome P450 can be found in biochemical literature. History Cytochromes were initially described in 1884 by Charles Alexander MacMunn as respiratory pigments (myohematin or histohematin). In the 1920s, Keilin rediscovered these respiratory pigments and named them the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rieske Protein
Rieske proteins are iron–sulfur protein (ISP) components of cytochrome ''bc''1 complexes and cytochrome b6f complexes and are responsible for electron transfer in some biological systems. John S. Rieske and co-workers first discovered the protein and in 1964 isolated an acetylated form of the bovine mitochondrial protein. In 1979 Trumpower's lab isolated the "oxidation factor" from bovine mitochondria and showed it was a reconstitutively-active form of the Rieske iron-sulfur protein It is a unique Fe-2Scluster in that one of the two Fe atoms is coordinated by two histidine residues rather than two cysteine residues. They have since been found in plants, animals, and bacteria with widely ranging electron reduction potentials from -150 to +400 mV. Biological function Ubiquinol-cytochrome-c reductase (also known as bc1 complex or complex III) is an enzyme complex of bacterial and mitochondrial oxidative phosphorylation systems. It catalyses the oxidation-reduction reaction ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Iron-sulfur Protein
Iron–sulfur proteins (or iron–sulphur proteins in British spelling) are proteins characterized by the presence of iron–sulfur clusters containing sulfide-linked di-, tri-, and tetrairon centers in variable oxidation states. Iron–sulfur clusters are found in a variety of metalloproteins, such as the ferredoxins, as well as NADH dehydrogenase, hydrogenases, coenzyme Q – cytochrome c reductase, succinate – coenzyme Q reductase and nitrogenase. Iron–sulfur clusters are best known for their role in the oxidation-reduction reactions of electron transport in mitochondria and chloroplasts. Both Complex I and Complex II of oxidative phosphorylation have multiple Fe–S clusters. They have many other functions including catalysis as illustrated by aconitase, generation of radicals as illustrated by SAM-dependent enzymes, and as sulfur donors in the biosynthesis of lipoic acid and biotin. Additionally, some Fe–S proteins regulate gene expression. Fe–S proteins are vulner ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molybdenum
Molybdenum is a chemical element with the symbol Mo and atomic number 42 which is located in period 5 and group 6. The name is from Neo-Latin ''molybdaenum'', which is based on Ancient Greek ', meaning lead, since its ores were confused with lead ores. Molybdenum minerals have been known throughout history, but the element was discovered (in the sense of differentiating it as a new entity from the mineral salts of other metals) in 1778 by Carl Wilhelm Scheele. The metal was first isolated in 1781 by Peter Jacob Hjelm. Molybdenum does not occur naturally as a free metal on Earth; it is found only in various oxidation states in minerals. The free element, a silvery metal with a grey cast, has the sixth-highest melting point of any element. It readily forms hard, stable carbides in alloys, and for this reason most of the world production of the element (about 80%) is used in steel alloys, including high-strength alloys and superalloys. Most molybdenum compounds have low solubility ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
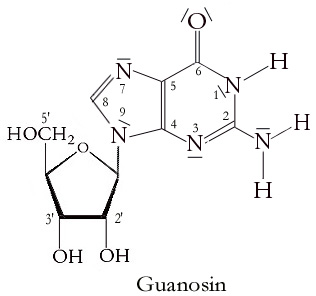
Guanosine Dinucleotide
Guanosine ( symbol G or Guo) is a purine nucleoside comprising guanine attached to a ribose (ribofuranose) ring via a β-N9-glycosidic bond. Guanosine can be phosphorylated to become guanosine monophosphate (GMP), cyclic guanosine monophosphate (cGMP), guanosine diphosphate (GDP), and guanosine triphosphate (GTP). These forms play important roles in various biochemical processes such as synthesis of nucleic acids and proteins, photosynthesis, muscle contraction, and intracellular signal transduction (cGMP). When guanine is attached by its N9 nitrogen to the C1 carbon of a deoxyribose ring it is known as deoxyguanosine. Physical and chemical properties Guanosine is a white, crystalline powder with no odor and mild saline taste. It is very soluble in acetic acid, slightly soluble in water, insoluble in ethanol, diethyl ether, benzene and chloroform. Functions Guanosine is required for an RNA splicing reaction in mRNA, when a "self-splicing" intron removes itself from the mRNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molybdopterin
Molybdopterins are a class of cofactors found in most molybdenum-containing and all tungsten-containing enzymes. Synonyms for molybdopterin are: MPT and pyranopterin-dithiolate. The nomenclature for this biomolecule can be confusing: Molybdopterin itself contains no molybdenum; rather, this is the name of the ligand (a '' pterin'') that will bind the active metal. After molybdopterin is eventually complexed with molybdenum, the complete ligand is usually called molybdenum cofactor. Molybdopterin consists of a pyranopterin, a complex heterocycle featuring a pyran fused to a pterin ring. In addition, the pyran ring features two thiolates, which serve as ligands in molybdo- and tungstoenzymes. In some cases, the alkyl phosphate group is replaced by an alkyl diphosphate nucleotide. Enzymes that contain the molybdopterin cofactor include xanthine oxidase, DMSO reductase, sulfite oxidase, and nitrate reductase. The only molybdenum-containing enzymes that do not feature molybdo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Enzymes
This article lists enzymes by their classification in the International Union of Biochemistry and Molecular Biology's Enzyme Commission (EC) numbering system. * List of EC numbers (EC 5) * List of EC numbers (EC 6) :Oxidoreductases (EC 1) (Oxidoreductase) * Dehydrogenase * Luciferase * DMSO reductase :EC 1.1 (act on the CH-OH group of donors) * :EC 1.1.1 (with NAD+ or NADP+ as acceptor) ** Alcohol dehydrogenase (NAD) ** Alcohol dehydrogenase (NADP) ** Homoserine dehydrogenase ** Aminopropanol oxidoreductase ** Diacetyl reductase **Glycerol dehydrogenase ** Propanediol-phosphate dehydrogenase ** glycerol-3-phosphate dehydrogenase (NAD+) ** D-xylulose reductase **L-xylulose reductase ** Lactate dehydrogenase ** Malate dehydrogenase ** Isocitrate dehydrogenase **HMG-CoA reductase * :EC 1.1.2 (with a cytochrome as acceptor) * :EC 1.1.3 (with oxygen as acceptor) ** Glucose oxidase ** L-gulonolactone oxidase ** Thiamine oxidase ** Xanthine oxidase * :EC 1.1.4 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oxidoreductase
In biochemistry, an oxidoreductase is an enzyme that catalyzes the transfer of electrons from one molecule, the reductant, also called the electron donor, to another, the oxidant, also called the electron acceptor. This group of enzymes usually utilizes NADP+ or NAD+ as cofactors. Transmembrane oxidoreductases create electron transport chains in bacteria, chloroplasts and mitochondria, including respiratory complexes I, II and III. Some others can associate with biological membranes as peripheral membrane proteins or be anchored to the membranes through a single transmembrane helix. in Membranome database Reactions For exa ...[...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Catalysis
Catalysis () is the process of increasing the rate of a chemical reaction by adding a substance known as a catalyst (). Catalysts are not consumed in the reaction and remain unchanged after it. If the reaction is rapid and the catalyst recycles quickly, very small amounts of catalyst often suffice; mixing, surface area, and temperature are important factors in reaction rate. Catalysts generally react with one or more reactants to form intermediates that subsequently give the final reaction product, in the process of regenerating the catalyst. Catalysis may be classified as either homogeneous, whose components are dispersed in the same phase (usually gaseous or liquid) as the reactant, or heterogeneous, whose components are not in the same phase. Enzymes and other biocatalysts are often considered as a third category. Catalysis is ubiquitous in chemical industry of all kinds. Estimates are that 90% of all commercially produced chemical products involve catalysts at some s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrogen Ion
A hydrogen ion is created when a hydrogen atom loses or gains an electron. A positively charged hydrogen ion (or proton) can readily combine with other particles and therefore is only seen isolated when it is in a gaseous state or a nearly particle-free space. Due to its extremely high charge density of approximately 2×1010 times that of a sodium ion, the bare hydrogen ion cannot exist freely in solution as it readily hydrates, i.e., bonds quickly. The hydrogen ion is recommended by IUPAC as a general term for all ions of hydrogen and its isotopes. Depending on the charge of the ion, two different classes can be distinguished: positively charged ions and negatively charged ions. Cation (positively charged) A hydrogen atom is made up of a nucleus with charge +1, and a single electron. Therefore, the only positively charged ion possible has charge +1. It is noted H+. Depending on the isotope in question, the hydrogen cation has different names: * Hydron: general name refe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |