|
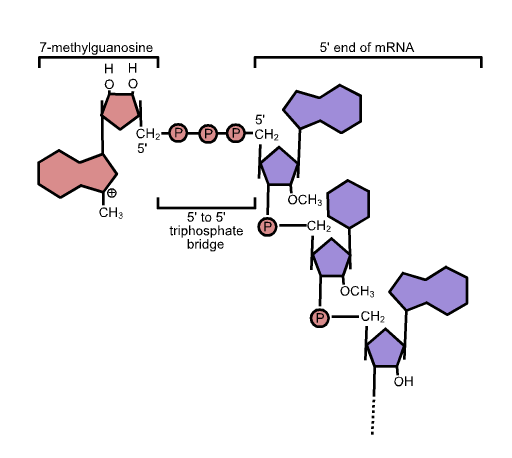
5′ Cap
In molecular biology, the five-prime cap (5′ cap) is a specially altered nucleotide on the 5′ end of some primary transcripts such as precursor messenger RNA. This process, known as mRNA capping, is highly regulated and vital in the creation of stable and mature messenger RNA able to undergo translation during protein synthesis. Mitochondrial mRNA and chloroplastic mRNA are not capped. Structure In eukaryotes, the 5′ cap (cap-0), found on the 5′ end of an mRNA molecule, consists of a guanine nucleotide connected to mRNA via an unusual 5′ to 5′ triphosphate linkage. This guanosine is methylated on the 7 position directly after capping ''in vivo'' by a methyltransferase. It is referred to as a 7-methylguanylate cap, abbreviated m7G. In multicellular eukaryotes and some viruses, further modifications exist, including the methylation of the 2′ hydroxy-groups of the first 2 ribose sugars of the 5′ end of the mRNA. cap-1 has a methylated 2′-hydroxy group ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Biology
Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions. The study of chemical and physical structure of biological macromolecules is known as molecular biology. Molecular biology was first described as an approach focused on the underpinnings of biological phenomena - uncovering the structures of biological molecules as well as their interactions, and how these interactions explain observations of classical biology. In 1945 the term molecular biology was used by physicist William Astbury. In 1953 Francis Crick, James Watson, Rosalind Franklin, and colleagues, working at Medical Research Council unit, Cavendish laboratory, Cambridge (now the MRC Laboratory of Molecular Biology), made a double helix model of DNA which changed the entire research scenario. They proposed the DNA structure based on previous research done by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methyltransferase
Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding ''S''-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the le ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Guanosine Triphosphate
Guanosine-5'-triphosphate (GTP) is a purine nucleoside triphosphate. It is one of the building blocks needed for the synthesis of RNA during the transcription process. Its structure is similar to that of the guanosine nucleoside, the only difference being that nucleotides like GTP have phosphates on their ribose sugar. GTP has the guanine nucleobase attached to the 1' carbon of the ribose and it has the triphosphate moiety attached to ribose's 5' carbon. It also has the role of a source of energy or an activator of substrates in metabolic reactions, like that of ATP, but more specific. It is used as a source of energy for protein synthesis and gluconeogenesis. GTP is essential to signal transduction, in particular with G-proteins, in second-messenger mechanisms where it is converted to guanosine diphosphate (GDP) through the action of GTPases. Uses Energy transfer GTP is involved in energy transfer within the cell. For instance, a GTP molecule is generated by o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Triphosphatase
The enzyme polynucleotide 5′-phosphatase (RNA 5′-triphosphatase, RTPase, EC 3.1.3.33) is an enzyme that catalyzes the reaction :a 5′-phosphopolynucleotide + H2O \rightleftharpoons a polynucleotide + phosphate This enzyme belongs to the family of hydrolases, specifically those acting on phosphoric monoester bonds. The systematic name is polynucleotide 5′-phosphohydrolase. This enzyme is also called 5′-polynucleotidase. The only specific molecular function known is the catalysis of the reaction: :a 5′-end triphospho-(purine-ribonucleotide) in mRNA + H2O = a 5′-end diphospho-(purine-ribonucleoside) in mRNA + phosphate RTPases cleave the 5′-terminal γ-β phosphoanhydride bond of nascent messenger RNA molecules, enabling the addition of a five-prime cap as part of post-transcriptional modifications. RTPases generate 5′-diphosphate-ended mRNA and a phosphate ion from 5′-triphosphate-ended precursor mRNA. mRNA guanylyltransferase then adds a backwards guanosine ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Messenger RNA Decapping
The process of messenger RNA decapping consists of hydrolysis of the 5' cap structure on the RNA exposing a 5' monophosphate. In eukaryotes, this 5' monophosphate is a substrate for the 5' exonuclease Xrn1 and the mRNA is quickly destroyed. There are many situations which may lead to the removal of the cap, some of which are discussed below. In prokaryotes, the initial mRNA transcript naturally possesses a 5'-triphosphate group after bacterial transcription; the enzyme RppH removes a pyrophosphate molecule from the 5' end, converting the 5'-triphosphate to a 5'-monophosphate, triggering mRNA degradation by ribonucleases. Translation and decay Inside cells, there is a balance between the processes of translation and mRNA decay. Messages which are being actively translated are bound by polysomes and the eukaryotic initiation factors eIF-4E and eIF-4G (in eukaryotes). This blocks access to the cap by the decapping enzyme DCP2 and protects the mRNA molecule. In nutrient-star ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Coenzyme A
Coenzyme A (CoA, SHCoA, CoASH) is a coenzyme, notable for its role in the synthesis and oxidation of fatty acids, and the oxidation of pyruvate in the citric acid cycle. All genomes sequenced to date encode enzymes that use coenzyme A as a substrate, and around 4% of cellular enzymes use it (or a thioester) as a substrate. In humans, CoA biosynthesis requires cysteine, pantothenate (vitamin B5), and adenosine triphosphate (ATP). In its acetyl form, coenzyme A is a highly versatile molecule, serving metabolic functions in both the anabolic and catabolic pathways. Acetyl-CoA is utilised in the post-translational regulation and allosteric regulation of pyruvate dehydrogenase and carboxylase to maintain and support the partition of pyruvate synthesis and degradation. Discovery of structure Coenzyme A was identified by Fritz Lipmann in 1946, who also later gave it its name. Its structure was determined during the early 1950s at the Lister Institute, London, together by L ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NADH
Nicotinamide adenine dinucleotide (NAD) is a coenzyme central to metabolism. Found in all living cells, NAD is called a dinucleotide because it consists of two nucleotides joined through their phosphate groups. One nucleotide contains an adenine nucleobase and the other nicotinamide. NAD exists in two forms: an oxidized and reduced form, abbreviated as NAD and NADH (H for hydrogen), respectively. In metabolism, nicotinamide adenine dinucleotide is involved in redox reactions, carrying electrons from one reaction to another. The cofactor is, therefore, found in two forms in cells: NAD is an oxidizing agent – it accepts electrons from other molecules and becomes reduced. This reaction, also with H+, forms NADH, which can then be used as a reducing agent to donate electrons. These electron transfer reactions are the main function of NAD. However, it is also used in other cellular processes, most notably as a substrate of enzymes in adding or removing chemical groups to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nicotinamide Adenine Dinucleotide
Nicotinamide adenine dinucleotide (NAD) is a coenzyme central to metabolism. Found in all living cells, NAD is called a dinucleotide because it consists of two nucleotides joined through their phosphate groups. One nucleotide contains an adenine nucleobase and the other nicotinamide. NAD exists in two forms: an oxidized and reduced form, abbreviated as NAD and NADH (H for hydrogen), respectively. In metabolism, nicotinamide adenine dinucleotide is involved in redox reactions, carrying electrons from one reaction to another. The cofactor is, therefore, found in two forms in cells: NAD is an oxidizing agent – it accepts electrons from other molecules and becomes reduced. This reaction, also with H+, forms NADH, which can then be used as a reducing agent to donate electrons. These electron transfer reactions are the main function of NAD. However, it is also used in other cellular processes, most notably as a substrate of enzymes in adding or removing chemical ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria are vital in many stages of the nutrient cycle by recycling nutrients such as the fixation of nitrogen from the atmosphere. The nutrient cycle includes the decomposition of dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy. Bacteria also live in symbiotic and parasitic re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Small Nuclear RNA
Small nuclear RNA (snRNA) is a class of small RNA molecules that are found within the splicing speckles and Cajal bodies of the cell nucleus in eukaryotic cells. The length of an average snRNA is approximately 150 nucleotides. They are transcribed by either RNA polymerase II or RNA polymerase III. Their primary function is in the processing of pre- messenger RNA ( hnRNA) in the nucleus. They have also been shown to aid in the regulation of transcription factors ( 7SK RNA) or RNA polymerase II (B2 RNA), and maintaining the telomeres. snRNA are always associated with a set of specific proteins, and the complexes are referred to as small nuclear ribonucleoproteins (snRNP, often pronounced "snurps"). Each snRNP particle is composed of a snRNA component and several snRNP-specific proteins (including Sm proteins, a family of nuclear proteins). The most common human snRNA components of these complexes are known, respectively, as: U1 spliceosomal RNA, U2 spliceosomal RNA, U4 splic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Exonuclease
Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is the endonuclease, which cleaves phosphodiester bonds in the middle (endo) of a polynucleotide chain. Eukaryotes and prokaryotes have three types of exonucleases involved in the normal turnover of mRNA: 5′ to 3′ exonuclease (Xrn1), which is a dependent decapping protein; 3′ to 5′ exonuclease, an independent protein; and poly(A)-specific 3′ to 5′ exonuclease. In both archaea and eukaryotes, one of the main routes of RNA degradation is performed by the multi-protein exosome complex, which consists largely of 3′ to 5′ exoribonucleases. Significance to polymerase RNA polymerase II is known to be in effect during transcriptional termination; it works with a 5' exonuclease (human gene Xrn2) to degrade the newly formed tra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
3′ End
Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. In a single strand of DNA or RNA, the chemical convention of naming carbon atoms in the nucleotide pentose-sugar-ring means that there will be a 5′ end (usually pronounced "five-prime end"), which frequently contains a phosphate group attached to the 5′ carbon of the ribose ring, and a 3′ end (usually pronounced "three-prime end"), which typically is unmodified from the ribose -OH substituent. In a DNA double helix, the strands run in opposite directions to permit base pairing between them, which is essential for replication or transcription of the encoded information. Nucleic acids can only be synthesized in vivo in the 5′-to-3′ direction, as the polymerases that assemble various types of new strands generally rely on the energy produced by breaking nucleoside triphosphate bonds to attach new nucleoside monophosphates to the 3′- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |