|
Small Nuclear RNA
Small nuclear RNA (snRNA) is a class of small RNA molecules that are found within the Cell nucleus#Splicing speckles, splicing speckles and Cajal body, Cajal bodies of the cell nucleus in eukaryotic cells. The length of an average snRNA is approximately 150 nucleotides. They are transcribed by either RNA polymerase II or RNA polymerase III. Their primary function is in the processing of pre-messenger RNA (hnRNA) in the nucleus. They have also been shown to aid in the regulation of transcription factors (7SK RNA) or RNA polymerase II (B2 RNA), and maintaining the telomeres. snRNA are always associated with a set of specific proteins, and the complexes are referred to as small nuclear ribonucleoproteins (snRNP, often pronounced "snurps"). Each snRNP particle is composed of a snRNA component and several snRNP-specific proteins (including LSm, Sm proteins, a family of nuclear proteins). The most common human snRNA components of these complexes are known, respectively, as: U1 spliceos ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Small RNA
Small RNA (sRNA) are polymeric RNA molecules that are less than 200 nucleotides in length, and are usually non-coding RNA, non-coding. RNA silencing is often a function of these molecules, with the most common and well-studied example being RNA interference (RNAi), in which endogenously (from within the organism) expressed microRNA (miRNA) or endogenously/exogenously (from outside the organism) derived small interfering RNA (siRNA) induces the degradation of complementarity (molecular biology), complementary messenger RNA. Other classes of small RNA have been identified, including piwi-interacting RNA (piRNA) and its subspecies rasiRNA, repeat associated small interfering RNA (rasiRNA). Small RNA "is unable to induce RNAi alone, and to accomplish the task it must form the core of the RNA–protein complex termed the RNA-induced silencing complex (RISC), specifically with Argonaute protein". Small RNA have been detected or sequenced using a range of techniques, including directly by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
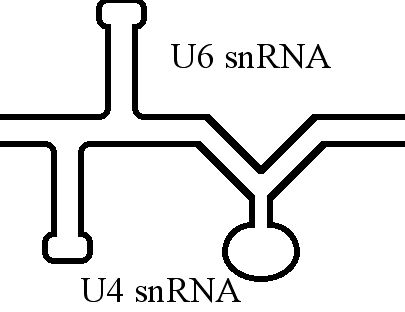
U6 Spliceosomal RNA
U6 snRNA is the non-coding small nuclear RNA (snRNA) component of U6 snRNP (''small nuclear ribonucleoprotein''), an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex that catalyzes the excision of introns from pre-mRNA. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification and takes place only in the nucleus of eukaryotes. The RNA sequence of U6 is the most highly conserved across species of all five of the snRNAs involved in the spliceosome, suggesting that the function of the U6 snRNA has remained both crucial and unchanged through evolution. It is common in vertebrate genomes to find many copies of the U6 snRNA gene or U6-derived pseudogenes. This prevalence of "back-ups" of the U6 snRNA gene in vertebrates further implies its evolutionary importance to organism viability. The U6 snRNA gene has been isolated in many org ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Survival Of Motor Neuron
Survival of motor neuron or survival motor neuron (SMN) is a protein that in humans is encoded by the ''SMN1'' and '' SMN2'' genes. SMN is found in the cytoplasm of all animal cells and also in the nuclear gems. It functions in transcriptional regulation, telomerase regeneration and cellular trafficking. SMN deficiency, primarily due to mutations in ''SMN1'', results in widespread splicing defects, especially in spinal motor neurons, and is one cause of spinal muscular atrophy. Research also showed a possible role of SMN in neuronal migration and/or differentiation. Function The SMN protein contains GEMIN2-binding, Tudor and YG-Box domains. It localizes to both the cytoplasm and the nucleus. Within the nucleus, the protein localizes to subnuclear bodies called gems which are found near coiled bodies containing high concentrations of small ribonucleoproteins (snRNPs). This protein forms heteromeric complexes with proteins such as GEMIN2 and GEMIN4, and also interacts w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Pores
The nuclear pore complex (NPC), is a large protein complex giving rise to the nuclear pore. A great number of nuclear pores are studded throughout the nuclear envelope that surrounds the eukaryote cell nucleus. The pores enable the nuclear transport of macromolecules between the nucleoplasm of the nucleus and the cytoplasm of the cell. Small molecules can easily diffuse through the pores. Nuclear transport includes the transportation of RNA and ribosomal proteins from the nucleus to the cytoplasm, and the transport of proteins (such as DNA polymerase and lamins), carbohydrates, signaling molecules, and lipids into the nucleus. Each nuclear pore complex can actively mediate up to 1000 translocations per second. The nuclear pore complex consists predominantly of a family of proteins known as nucleoporins (Nups). Each pore complex in the human cell nucleus is composed of about 1,000 individual protein molecules, from an evolutionarily conserved set of 35 distinct nucleoporins. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Five-prime Cap
In molecular biology, the five-prime cap (5′ cap) is a specially altered nucleotide on the 5′ end of some primary transcripts such as precursor messenger RNA. This process, known as mRNA capping, is highly regulated and vital in the creation of stable and mature messenger RNA able to undergo translation during protein synthesis. Mitochondrial mRNA and chloroplastic mRNA are not capped. Structure In eukaryotes, the 5′ cap (cap-0), found on the 5′ end of an mRNA molecule, consists of a guanine nucleotide connected to mRNA via an unusual 5′ to 5′ triphosphate linkage. This guanosine is methylated on the 7 position directly after capping ''in vivo'' by a methyltransferase. It is referred to as a 7-methylguanylate cap, abbreviated m7G. The Cap-0 is the base cap structure, however, the first and second transcribed nucleotides can also be 2' O-methylated, leading to the Cap-1 and Cap-2 structures, respectively. This is more common in higher eukaryotes an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
U12 Minor Spliceosomal RNA
U12 minor spliceosomal RNA is formed from U12 small nuclear ( snRNA), together with U4atac/ U6atac, U5, and U11 snRNAs and associated proteins, forms a spliceosome that cleaves a divergent class of low-abundance pre-mRNA introns. Although the U12 sequence is very divergent from that of U2, the two are functionally analogous. Structure The predicted secondary structure of U12 RNA is published,. However, the alternative single hairpin in the 3' end shown here seems to better match the alignment of divergent ''Drosophila melanogaster'' and ''Arabidopsis thaliana'' sequences. The sequences U12 introns that are spliced out are collected in a biological database Biological databases are libraries of biological sciences, collected from scientific experiments, published literature, high-throughput experiment technology, and computational analysis. They contain information from research areas including geno ... called the U12 intron database. References External links * ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
U11 Spliceosomal RNA
The U11 snRNA ( small nuclear ribonucleic acid) is an important non-coding RNA in the minor spliceosome protein complex, which activates the alternative splicing mechanism. The minor spliceosome is associated with similar protein components as the major spliceosome. It uses U11 snRNA to recognize the 5' splice site (functionally equivalent to U1 snRNA) while U12 snRNA binds to the branchpoint to recognize the 3' splice site (functionally equivalent to U2 snRNA). Secondary structure U11 snRNA has a stem-loop structure with a 5' end as splice site sequence (5' ss) and contains four stem loops structures (I-IV). A structural comparison of U11 snRNA between plants, vertebrates and insects shows that it is folded into a structure with a four-way junction at the 5' site and in a stem loop structure at the 3' site. Binding site during assembly pathway The 5' splice site region possesses sequence complementarity with the 5' splice site of the eukaryotic U12 type pre-mRNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
U7 Small Nuclear RNA
The U7 small nuclear RNA (U7 snRNA) is an RNA molecule and a component of the small nuclear ribonucleoprotein complex (U7 snRNP). The U7 snRNA is required for histone pre-mRNA processing. The 5' end of the U7 snRNA binds the HDE (histone downstream element), a conserved purine-rich region, located 15 nucleotides downstream the Histone 3' UTR stem-loop, histone mRNA cleavage site. The binding of the HDE region by the U7 snRNA, through complementary base-pairing, is an important step for the future recruitment of cleavage factors during histone pre-mRNA processing. See also *Duchenne muscular dystrophy *Histone 3' UTR stem-loop *LSM10 References Further reading External links * {{Rfam, id=RF00066, name=U7 small nuclear RNA The uRNA database RNA splicing Small nuclear RNA Spliceosome ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
U4atac Minor Spliceosomal RNA
U4atac minor spliceosomal RNA is a ncRNA which is an essential component of the minor U12-type spliceosome complex. The U12-type spliceosome is required for removal of the rarer class of eukaryotic introns (AT-AC, U12-type). U4atac snRNA is proposed to form a base-paired complex with another spliceosomal RNA U6atac via two stem loop regions. These interacting stem loops have been shown to be required for in vivo splicing. U4atac also contains a 3' Sm protein binding site which has been shown to be essential for splicing activity. U4atac is the functional analog of U4 spliceosomal RNA in the major U2-type spliceosomal complex. The Drosophila ''Drosophila'' (), from Ancient Greek δρόσος (''drósos''), meaning "dew", and φίλος (''phílos''), meaning "loving", is a genus of fly, belonging to the family Drosophilidae, whose members are often called "small fruit flies" or p ... U4atac snRNA has an additional predicted 3' stem loop terminal to the Sm binding si ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Small Cajal Body-specific RNA
Small Cajal body-specific RNAs (scaRNAs) are a class of small nucleolar RNAs (snoRNAs) that specifically localise to the Cajal body, a nuclear organelle (cellular sub-organelle) involved in the biogenesis of small nuclear ribonucleoproteins (snRNPs or snurps). ScaRNAs guide the modification (methylation and pseudouridylation) of RNA polymerase II transcribed spliceosomal RNAs U1, U2, U4, U5 and U12. The first scaRNA identified was U85. It is unlike typical snoRNAs in that it is a composite C/D box and H/ACA box snoRNAs and can guide both pseudouridylation and 2′-''O''-methylation. Not all scaRNAs are composite C/D and H/ACA box snoRNA and most scaRNAs are structurally and functionally indistinguishable from snoRNAs, directing ribosomal RNA (rRNA) modification in the nucleolus. ''Drosophila'' scaRNAs Several studies identified scaRNAs in ''Drosophila.'' One of the studies showed that several ''Drosophila'' scaRNAs could function equally well in the nucleoplasm of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleolus
The nucleolus (; : nucleoli ) is the largest structure in the cell nucleus, nucleus of eukaryote, eukaryotic cell (biology), cells. It is best known as the site of ribosome biogenesis. The nucleolus also participates in the formation of signal recognition particles and plays a role in the cell's response to stress. Nucleoli are made of proteins, DNA and RNA, and form around specific chromosomal regions called nucleolar organizing regions. Malfunction of the nucleolus is the cause of several human conditions called "nucleolopathies" and the nucleolus is being investigated as a target for cancer chemotherapy. History The nucleolus was identified by bright-field microscopy during the 1830s. Theodor Schwann in his 1839 treatise described that Matthias Jakob Schleiden, Schleiden had identified small corpuscles in nuclei, and named the structures "Kernkörperchen". In a 1947 translation of the work to English, the structure was named "nucleolus". Little was known about the fun ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biogenesis
Spontaneous generation is a Superseded scientific theories, superseded scientific theory that held that living creatures could arise from abiotic component, non-living matter and that such processes were commonplace and regular. It was Hypothesis, hypothesized that certain forms, such as fleas, could arise from inanimate matter such as dust, or that maggots could arise from dead flesh. The doctrine of spontaneous generation was coherently synthesized by the Greek philosopher and naturalist Aristotle, who compiled and expanded the work of Pre-Socratic philosophy, earlier natural philosophers and the various ancient explanations for the appearance of organisms. Spontaneous generation was taken as scientific fact for two millennia. Though challenged in the 17th and 18th centuries by the experiments of the Italian biologists Francesco Redi and Lazzaro Spallanzani, it was not discredited until the work of the French chemist Louis Pasteur and the Irish physicist John Tyndall in the m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |