Protein Structures on:
[Wikipedia]
[Google]
[Amazon]
Protein structure is the three-dimensional arrangement of atoms in an


 Proteins are frequently described as consisting of several structural units. These units include domains, motifs, and folds. Despite the fact that there are about 100,000 different proteins expressed in
Proteins are frequently described as consisting of several structural units. These units include domains, motifs, and folds. Despite the fact that there are about 100,000 different proteins expressed in
 Proteins are often thought of as relatively stable tertiary structures that experience conformational changes after being affected by interactions with other proteins or as a part of enzymatic activity. However, proteins may have varying degrees of stability, and some of the less stable variants are
Proteins are often thought of as relatively stable tertiary structures that experience conformational changes after being affected by interactions with other proteins or as a part of enzymatic activity. However, proteins may have varying degrees of stability, and some of the less stable variants are
Protein Ensemble Database
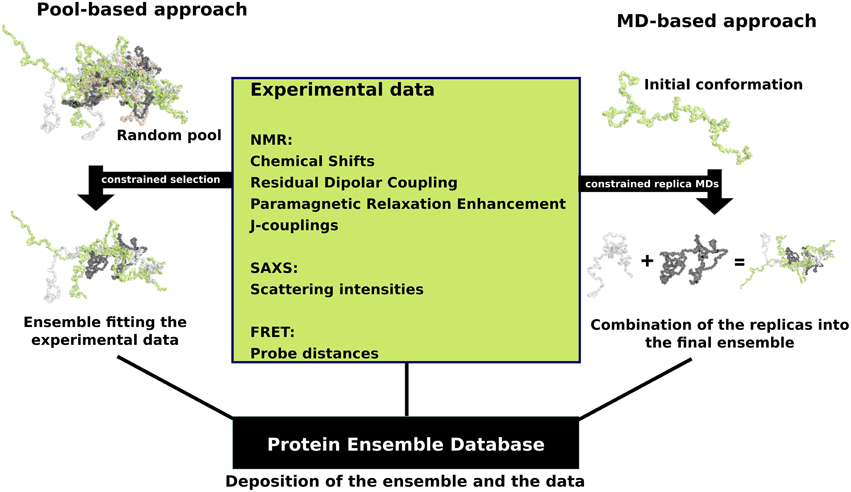
that fall into two general methodologies – pool and molecular dynamics (MD) approaches (diagrammed in the figure). The pool based approach uses the protein's amino acid sequence to create a massive pool of random conformations. This pool is then subjected to more computational processing that creates a set of theoretical parameters for each conformation based on the structure. Conformational subsets from this pool whose average theoretical parameters closely match known experimental data for this protein are selected. The alternative molecular dynamics approach takes multiple random conformations at a time and subjects all of them to experimental data. Here the experimental data is serving as limitations to be placed on the conformations (e.g. known distances between atoms). Only conformations that manage to remain within the limits set by the experimental data are accepted. This approach often applies large amounts of experimental data to the conformations which is a very computationally demanding task. The conformational ensembles were generated for a number of highly dynamic and partially unfolded proteins, such as
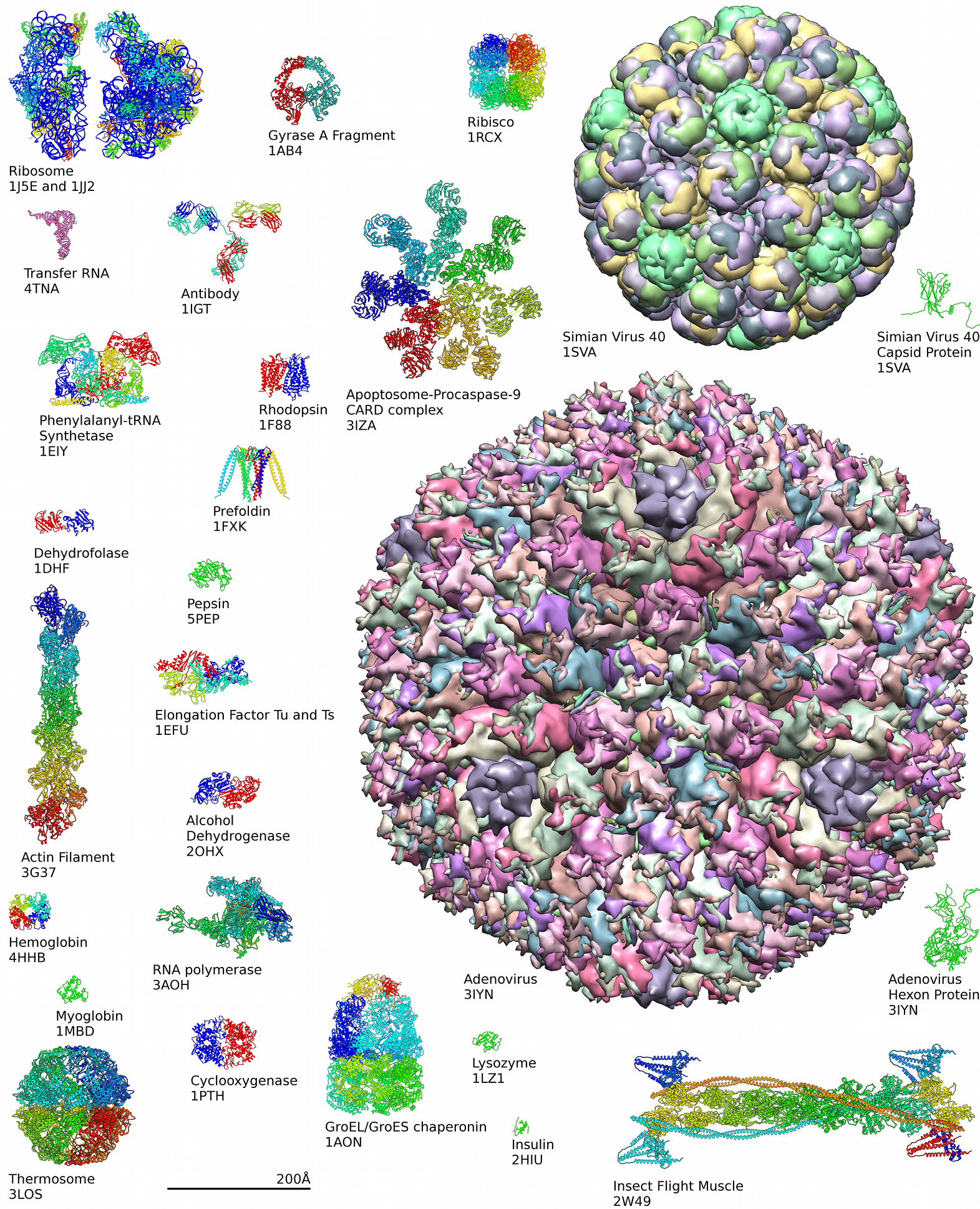
 Around 90% of the protein structures available in the
Around 90% of the protein structures available in the
50 Years of Protein Structure Determination Timeline - HTML Version - National Institute of General Medical Sciences
at
Protein Structure
drugdesign.org
Method_for_the_Characterization_of_the_Three-Dimensional_Structure_of_Proteins_Employing_Mass_Spectrometric_Analysis_and_Experimental-Computational_Feedback_Modeling
A_Method_for_the_Determination_of_the_Conformation_(Topology)_of_Proteins_Employing_Experimental-Computational_Feedback_Modeling
amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
-chain molecule
A molecule is a group of two or more atoms that are held together by Force, attractive forces known as chemical bonds; depending on context, the term may or may not include ions that satisfy this criterion. In quantum physics, organic chemi ...
. Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
s are polymer
A polymer () is a chemical substance, substance or material that consists of very large molecules, or macromolecules, that are constituted by many repeat unit, repeating subunits derived from one or more species of monomers. Due to their br ...
s specifically polypeptide
Peptides are short chains of amino acids linked by peptide bonds. A polypeptide is a longer, continuous, unbranched peptide chain. Polypeptides that have a molecular mass of 10,000 Da or more are called proteins. Chains of fewer than twenty ...
s formed from sequences of amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
s, which are the monomer
A monomer ( ; ''mono-'', "one" + '' -mer'', "part") is a molecule that can react together with other monomer molecules to form a larger polymer chain or two- or three-dimensional network in a process called polymerization.
Classification
Chemis ...
s of the polymer. A single amino acid monomer may also be called a ''residue'', which indicates a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reaction
In organic chemistry, a condensation reaction is a type of chemical reaction in which two molecules are combined to form a single molecule, usually with the loss of a small molecule such as water. If water is lost, the reaction is also known as a ...
s, in which the amino acids lose one water molecule
Water () is a polar inorganic compound that is at room temperature a tasteless and odorless liquid, which is nearly colorless apart from an inherent hint of blue. It is by far the most studied chemical compound and is described as the "univ ...
per reaction
Reaction may refer to a process or to a response to an action, event, or exposure.
Physics and chemistry
*Chemical reaction
*Nuclear reaction
*Reaction (physics), as defined by Newton's third law
* Chain reaction (disambiguation)
Biology and ...
in order to attach to one another with a peptide bond
In organic chemistry, a peptide bond is an amide type of covalent chemical bond linking two consecutive alpha-amino acids from C1 (carbon number one) of one alpha-amino acid and N2 (nitrogen number two) of another, along a peptide or protein cha ...
. By convention, a chain under 30 amino acids is often identified as a peptide
Peptides are short chains of amino acids linked by peptide bonds. A polypeptide is a longer, continuous, unbranched peptide chain. Polypeptides that have a molecular mass of 10,000 Da or more are called proteins. Chains of fewer than twenty am ...
, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions, such as hydrogen bonding
In chemistry, a hydrogen bond (H-bond) is a specific type of molecular interaction that exhibits partial covalent character and cannot be described as a purely electrostatic force. It occurs when a hydrogen (H) atom, Covalent bond, covalently b ...
, ionic interaction
Ionic bonding is a type of chemical bonding that involves the electrostatic attraction between oppositely charged ions, or between two atoms with sharply different electronegativities, and is the primary interaction occurring in ionic compound ...
s, Van der Waals forces
In molecular physics and chemistry, the van der Waals force (sometimes van der Waals' force) is a distance-dependent interaction between atoms or molecules. Unlike ionic or covalent bonds, these attractions do not result from a chemical ele ...
, and hydrophobic
In chemistry, hydrophobicity is the chemical property of a molecule (called a hydrophobe) that is seemingly repelled from a mass of water. In contrast, hydrophiles are attracted to water.
Hydrophobic molecules tend to be nonpolar and, thu ...
packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their three-dimensional structure. This is the topic of the scientific field of structural biology
Structural biology deals with structural analysis of living material (formed, composed of, and/or maintained and refined by living cells) at every level of organization.
Early structural biologists throughout the 19th and early 20th centuries we ...
, which employs techniques such as X-ray crystallography
X-ray crystallography is the experimental science of determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to Diffraction, diffract in specific directions. By measuring th ...
, NMR spectroscopy
Nuclear magnetic resonance spectroscopy, most commonly known as NMR spectroscopy or magnetic resonance spectroscopy (MRS), is a spectroscopic technique based on re-orientation of atomic nuclei with non-zero nuclear spins in an external magnetic f ...
, cryo-electron microscopy (cryo-EM) and dual polarisation interferometry
Dual-polarization interferometry (DPI) is an analytical technique that probes molecular layers adsorbed to the surface of a waveguide using the evanescent wave of a laser beam. It is used to measure the conformational change in proteins, or o ...
, to determine the structure of proteins.
Protein structures range in size from tens to several thousand amino acids. By physical size, proteins are classified as nanoparticle
A nanoparticle or ultrafine particle is a particle of matter 1 to 100 nanometres (nm) in diameter. The term is sometimes used for larger particles, up to 500 nm, or fibers and tubes that are less than 100 nm in only two directions. At ...
s, between 1–100 nm. Very large protein complexes
A protein complex or multiprotein complex is a group of two or more associated polypeptide chains. Protein complexes are distinct from multidomain enzymes, in which multiple catalytic domains are found in a single polypeptide chain.
Protein c ...
can be formed from protein subunit
In structural biology, a protein subunit is a polypeptide chain or single protein molecule that assembles (or "''coassembles''") with others to form a protein complex.
Large assemblies of proteins such as viruses often use a small number of t ...
s. For example, many thousands of actin
Actin is a family of globular multi-functional proteins that form microfilaments in the cytoskeleton, and the thin filaments in muscle fibrils. It is found in essentially all eukaryotic cells, where it may be present at a concentration of ...
molecules assemble into a microfilament
Microfilaments, also called actin filaments, are protein filaments in the cytoplasm of eukaryotic cells that form part of the cytoskeleton. They are primarily composed of polymers of actin, but are modified by and interact with numerous other ...
.
A protein usually undergoes reversible structural changes in performing its biological function. The alternative structures of the same protein are referred to as different conformations, and transitions between them are called conformational change
In biochemistry, a conformational change is a change in the shape of a macromolecule, often induced by environmental factors.
A macromolecule is usually flexible and dynamic. Its shape can change in response to changes in its environment or othe ...
s.
Levels of protein structure
There are four distinct levels of protein structure.
Primary structure
Theprimary structure
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthe ...
of a protein refers to the sequence of amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
s in the polypeptide chain. The primary structure is held together by peptide bonds
In organic chemistry, a peptide bond is an amide type of covalent chemical bond linking two consecutive alpha-amino acids from C1 (carbon number one) of one alpha-amino acid and N2 (nitrogen number two) of another, along a peptide or protein chai ...
that are made during the process of protein biosynthesis
Protein biosynthesis, or protein synthesis, is a core biological process, occurring inside Cell (biology), cells, homeostasis, balancing the loss of cellular proteins (via Proteolysis, degradation or Protein targeting, export) through the produc ...
. The two ends of the polypeptide chain
Peptides are short chains of amino acids linked by peptide bonds. A polypeptide is a longer, continuous, unbranched peptide chain. Polypeptides that have a molecular mass of 10,000 Da or more are called proteins. Chains of fewer than twenty ami ...
are referred to as the carboxyl terminus (C-terminus) and the amino terminus
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amin ...
(N-terminus) based on the nature of the free group on each extremity. Counting of residues always starts at the N-terminal end (NH2-group), which is the end where the amino group is not involved in a peptide bond. The primary structure of a protein is determined by the gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei ...
corresponding to the protein. A specific sequence of nucleotide
Nucleotides are Organic compound, organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both o ...
s in DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
is transcribed into mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein.
mRNA is ...
, which is read by the ribosome
Ribosomes () are molecular machine, macromolecular machines, found within all cell (biology), cells, that perform Translation (biology), biological protein synthesis (messenger RNA translation). Ribosomes link amino acids together in the order s ...
in a process called translation
Translation is the communication of the semantics, meaning of a #Source and target languages, source-language text by means of an Dynamic and formal equivalence, equivalent #Source and target languages, target-language text. The English la ...
. The sequence of amino acids in insulin was discovered by Frederick Sanger
Frederick Sanger (; 13 August 1918 – 19 November 2013) was a British biochemist who received the Nobel Prize in Chemistry twice.
He won the 1958 Chemistry Prize for determining the amino acid sequence of insulin and numerous other prote ...
, establishing that proteins have defining amino acid sequences. The sequence of a protein is unique to that protein, and defines the structure and function of the protein. The sequence of a protein can be determined by methods such as Edman degradation or tandem mass spectrometry
Tandem mass spectrometry, also known as MS/MS or MS2, is a technique in instrumental analysis where two or more stages of analysis using one or more mass analyzer are performed with an additional reaction step in between these analyses to increa ...
. Often, however, it is read directly from the sequence of the gene using the genetic code
Genetic code is a set of rules used by living cell (biology), cells to Translation (biology), translate information encoded within genetic material (DNA or RNA sequences of nucleotide triplets or codons) into proteins. Translation is accomplished ...
. It is strictly recommended to use the words "amino acid residues" when discussing proteins because when a peptide bond is formed, a water molecule
Water () is a polar inorganic compound that is at room temperature a tasteless and odorless liquid, which is nearly colorless apart from an inherent hint of blue. It is by far the most studied chemical compound and is described as the "univ ...
is lost, and therefore proteins are made up of amino acid residues. Post-translational modification
In molecular biology, post-translational modification (PTM) is the covalent process of changing proteins following protein biosynthesis. PTMs may involve enzymes or occur spontaneously. Proteins are created by ribosomes, which translation (biolog ...
s such as phosphorylation
In biochemistry, phosphorylation is described as the "transfer of a phosphate group" from a donor to an acceptor. A common phosphorylating agent (phosphate donor) is ATP and a common family of acceptor are alcohols:
:
This equation can be writ ...
s and glycosylation
Glycosylation is the reaction in which a carbohydrate (or ' glycan'), i.e. a glycosyl donor, is attached to a hydroxyl or other functional group of another molecule (a glycosyl acceptor) in order to form a glycoconjugate. In biology (but not ...
s are usually also considered a part of the primary structure, and cannot be read from the gene. For example, insulin
Insulin (, from Latin ''insula'', 'island') is a peptide hormone produced by beta cells of the pancreatic islets encoded in humans by the insulin (''INS)'' gene. It is the main Anabolism, anabolic hormone of the body. It regulates the metabol ...
is composed of 51 amino acids in 2 chains. One chain has 31 amino acids, and the other has 20 amino acids.
Secondary structure

Secondary structure
Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta ...
refers to highly regular local sub-structures on the actual polypeptide backbone chain. Two main types of secondary structure, the α-helix
An alpha helix (or α-helix) is a sequence of amino acids in a protein that are twisted into a coil (a helix).
The alpha helix is the most common structural arrangement in the Protein secondary structure, secondary structure of proteins. It is al ...
and the β-strand
The beta sheet (β-sheet, also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a gene ...
or β-sheet
The beta sheet (β-sheet, also β-pleated sheet) is a common structural motif, motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone chain, backbon ...
s, were suggested in 1951 by Linus Pauling
Linus Carl Pauling ( ; February 28, 1901August 19, 1994) was an American chemist and peace activist. He published more than 1,200 papers and books, of which about 850 dealt with scientific topics. ''New Scientist'' called him one of the 20 gre ...
. These secondary structures are defined by patterns of hydrogen bonds
In chemistry, a hydrogen bond (H-bond) is a specific type of molecular interaction that exhibits partial covalent character and cannot be described as a purely electrostatic force. It occurs when a hydrogen (H) atom, covalently bonded to a mo ...
between the main-chain peptide groups. They have a regular geometry, being constrained to specific values of the dihedral angles ψ and φ on the Ramachandran plot
In biochemistry, a Ramachandran plot (also known as a Rama plot, a Ramachandran diagram or a �,ψplot), originally developed in 1963 by G. N. Ramachandran, C. Ramakrishnan, and V. Sasisekharan, is a way to visualize energetically allowed regio ...
. Both the α-helix and the β-sheet represent a way of saturating all the hydrogen bond donors and acceptors in the peptide backbone. Some parts of the protein are ordered but do not form any regular structures. They should not be confused with random coil
In polymer chemistry, a random coil is a conformation of polymers where the monomer subunits are oriented randomly while still being bonded to adjacent units. It is not one specific shape, but a statistical distribution of shapes for all the cha ...
, an unfolded polypeptide chain lacking any fixed three-dimensional structure. Several sequential secondary structures may form a " supersecondary unit".
Tertiary structure
Tertiary structure
Protein tertiary structure is the three-dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains and the ...
refers to the three-dimensional structure created by a single protein molecule (a single polypeptide chain
Peptides are short chains of amino acids linked by peptide bonds. A polypeptide is a longer, continuous, unbranched peptide chain. Polypeptides that have a molecular mass of 10,000 Da or more are called proteins. Chains of fewer than twenty ami ...
). It may include one or several domains. The α-helices and β-pleated-sheets are folded into a compact globular structure. The folding is driven by the ''non-specific'' hydrophobic interactions
The hydrophobic effect is the observed tendency of nonpolar substances to aggregate in an aqueous solution and to be excluded by water. The word hydrophobic literally means "water-fearing", and it describes the segregation of water and nonpolar ...
, the burial of hydrophobic residues
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 ap ...
from water
Water is an inorganic compound with the chemical formula . It is a transparent, tasteless, odorless, and Color of water, nearly colorless chemical substance. It is the main constituent of Earth's hydrosphere and the fluids of all known liv ...
, but the structure is stable only when the parts of a protein domain
In molecular biology, a protein domain is a region of a protein's Peptide, polypeptide chain that is self-stabilizing and that Protein folding, folds independently from the rest. Each domain forms a compact folded Protein tertiary structure, thre ...
are locked into place by ''specific'' tertiary interactions, such as salt bridges, hydrogen bonds, and the tight packing of side chains and disulfide bond
In chemistry, a disulfide (or disulphide in British English) is a compound containing a functional group or the anion. The linkage is also called an SS-bond or sometimes a disulfide bridge and usually derived from two thiol groups.
In inor ...
s. The disulfide bonds are extremely rare in cytosolic proteins, since the cytosol
The cytosol, also known as cytoplasmic matrix or groundplasm, is one of the liquids found inside cells ( intracellular fluid (ICF)). It is separated into compartments by membranes. For example, the mitochondrial matrix separates the mitochondri ...
(intracellular fluid) is generally a reducing environment.
Quaternary structure
Quaternary structure is the three-dimensional structure consisting of the aggregation of two or more individual polypeptide chains (subunits) that operate as a single functional unit ( multimer). The resulting multimer is stabilized by the same non-covalent interactions and disulfide bonds as in tertiary structure. There are many possible quaternary structure organisations. Complexes of two or more polypeptides (i.e. multiple subunits) are called multimers. Specifically it would be called a dimer if it contains two subunits, a trimer if it contains three subunits, atetramer
A tetramer () (''tetra-'', "four" + '' -mer'', "parts") is an oligomer formed from four monomers or subunits. The associated property is called ''tetramery''. An example from inorganic chemistry is titanium methoxide with the empirical formula ...
if it contains four subunits, and a pentamer
A pentamer is an entity composed of five subunits.
In chemistry, it applies to molecules made of five monomers.
In biochemistry, it applies to macromolecules, particularly pentameric proteins, made of five protein sub-units.
In microbiology, a ...
if it contains five subunits, and so forth. The subunits are frequently related to one another by symmetry operations, such as a 2-fold axis in a dimer. Multimers made up of identical subunits are referred to with a prefix of "homo-" and those made up of different subunits are referred to with a prefix of "hetero-", for example, a heterotetramer, such as the two alpha and two beta chains of hemoglobin
Hemoglobin (haemoglobin, Hb or Hgb) is a protein containing iron that facilitates the transportation of oxygen in red blood cells. Almost all vertebrates contain hemoglobin, with the sole exception of the fish family Channichthyidae. Hemoglobin ...
.
Homomers
An assemblage of multiple copies of a particularpolypeptide
Peptides are short chains of amino acids linked by peptide bonds. A polypeptide is a longer, continuous, unbranched peptide chain. Polypeptides that have a molecular mass of 10,000 Da or more are called proteins. Chains of fewer than twenty ...
chain can be described as a homomer, multimer or oligomer
In chemistry and biochemistry, an oligomer () is a molecule that consists of a few repeating units which could be derived, actually or conceptually, from smaller molecules, monomers.Quote: ''Oligomer molecule: A molecule of intermediate relativ ...
. Bertolini et al. in 2021 presented evidence that homomer formation may be driven by interaction between nascent polypeptide chains as they are translated from mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein.
mRNA is ...
by nearby adjacent ribosome
Ribosomes () are molecular machine, macromolecular machines, found within all cell (biology), cells, that perform Translation (biology), biological protein synthesis (messenger RNA translation). Ribosomes link amino acids together in the order s ...
s. Hundreds of proteins have been identified as being assembled into homomers in human cells. The process of assembly is often initiated by the interaction of the N-terminal region of polypeptide chains. Evidence that numerous gene products form homomers (multimers) in a variety of organisms based on intragenic complementation evidence was reviewed in 1965.
Domains, motifs, and folds in protein structure
 Proteins are frequently described as consisting of several structural units. These units include domains, motifs, and folds. Despite the fact that there are about 100,000 different proteins expressed in
Proteins are frequently described as consisting of several structural units. These units include domains, motifs, and folds. Despite the fact that there are about 100,000 different proteins expressed in eukaryotic
The eukaryotes ( ) constitute the Domain (biology), domain of Eukaryota or Eukarya, organisms whose Cell (biology), cells have a membrane-bound cell nucleus, nucleus. All animals, plants, Fungus, fungi, seaweeds, and many unicellular organisms ...
systems, there are many fewer different domains, structural motifs and folds.
Structural domain
Astructural domain
In molecular biology, a protein domain is a region of a protein's Peptide, polypeptide chain that is self-stabilizing and that Protein folding, folds independently from the rest. Each domain forms a compact folded Protein tertiary structure, thre ...
is an element of the protein's overall structure that is self-stabilizing and often folds independently of the rest of the protein chain. Many domains are not unique to the protein products of one gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei ...
or one gene family
A gene family is a set of several similar genes, formed by duplication of a single original gene, and generally with similar biochemical functions. One such family are the genes for human hemoglobin subunits; the ten genes are in two clusters on ...
but instead appear in a variety of proteins. Domains often are named and singled out because they figure prominently in the biological function of the protein they belong to; for example, the "calcium
Calcium is a chemical element; it has symbol Ca and atomic number 20. As an alkaline earth metal, calcium is a reactive metal that forms a dark oxide-nitride layer when exposed to air. Its physical and chemical properties are most similar to it ...
-binding domain of calmodulin
Calmodulin (CaM) (an abbreviation for calcium-modulated protein) is a multifunctional intermediate calcium-binding messenger protein expressed in all Eukaryote, eukaryotic cells. It is an intracellular target of the Second messenger system, sec ...
". Because they are independently stable, domains can be "swapped" by genetic engineering
Genetic engineering, also called genetic modification or genetic manipulation, is the modification and manipulation of an organism's genes using technology. It is a set of Genetic engineering techniques, technologies used to change the genet ...
between one protein and another to make chimera
Chimera, Chimaera, or Chimaira (Greek for " she-goat") originally referred to:
* Chimera (mythology), a fire-breathing monster of ancient Lycia said to combine parts from multiple animals
* Mount Chimaera, a fire-spewing region of Lycia or Cilicia ...
proteins. A conservative combination of several domains that occur in different proteins, such as protein tyrosine phosphatase
Protein tyrosine phosphatases (EC 3.1.3.48, systematic name protein-tyrosine-phosphate phosphohydrolase) are a group of enzymes that remove phosphate groups from phosphorylated tyrosine residues on proteins:
: proteintyrosine phosphate + H2O = ...
domain and C2 domain
A C2 domain is a protein structural domain involved in targeting proteins to cell membranes. The typical version (PKC-C2) has a beta-sandwich composed of 8 β-strands that co-ordinates two or three calcium ions, which bind in a cavity formed by th ...
pair, was called "a superdomain" that may evolve as a single unit.
Structural and sequence motifs
Thestructural
A structure is an arrangement and organization of interrelated elements in a material object or system, or the object or system so organized. Material structures include man-made objects such as buildings and machines and natural objects such as ...
and sequence motif
In biology, a sequence motif is a nucleotide or amino-acid sequence pattern that is widespread and usually assumed to be related to biological function of the macromolecule. For example, an ''N''-glycosylation site motif can be defined as ''A ...
s refer to short segments of protein three-dimensional structure or amino acid sequence that were found in a large number of different proteins
Supersecondary structure
Tertiary protein structures can have multiple secondary elements on the same polypeptide chain. Thesupersecondary structure
A supersecondary structure is a compact three-dimensional protein structure of several adjacent elements of a secondary structure that is smaller than a protein domain or a subunit. Supersecondary structures can act as nucleations in the process ...
refers to a specific combination of secondary structure
Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta ...
elements, such as β-α-β units or a helix-turn-helix
Helix-turn-helix is a DNA-binding domain (DBD). The helix-turn-helix (HTH) is a major structural motif capable of binding DNA. Each monomer incorporates two alpha helix, α helices, joined by a short strand of amino acids, that bind to the majo ...
motif. Some of them may be also referred to as structural motifs.
Protein fold
A protein fold refers to the general protein architecture, like ahelix bundle A helix bundle is a small protein fold composed of several alpha helices that are usually nearly parallel or antiparallel to each other.
Three-helix bundles
Three-helix bundles are among the smallest and fastest known cooperatively folding struct ...
, β-barrel
In protein structures, a beta barrel (β barrel) is a beta sheet (β sheet) composed of Protein tandem repeats, tandem repeats that twists and coils to form a closed toroidal structure in which the first strand is bonded to the last strand (hydrog ...
, Rossmann fold
The Rossmann fold is a tertiary fold found in proteins that bind nucleotides, such as enzyme cofactors FAD, NAD+, and NADP+. This fold is composed of alternating beta strands and alpha helical segments where the beta strands are hydrogen bond ...
or different "folds" provided in the Structural Classification of Proteins database
The Structural Classification of Proteins (SCOP) database is a largely manual classification of protein structural domains based on similarities of their structures and amino acid sequences. A motivation for this classification is to determine t ...
. A related concept is protein topology
Protein topology is a property of protein molecule that does not change under deformation (without cutting or breaking a bond). Frameworks
Two main topology frameworks have been developed and applied to protein molecules. Knot Theory
Knot theory ...
.
Protein dynamics and conformational ensembles
Proteins are not static objects, but rather populate ensembles of conformational states. Transitions between these states typically occur onnanoscale
Nanotechnology is the manipulation of matter with at least one dimension sized from 1 to 100 nanometers (nm). At this scale, commonly known as the nanoscale, surface area and quantum mechanical effects become important in describing propertie ...
s, and have been linked to functionally relevant phenomena such as allosteric signaling and enzyme catalysis
Enzyme catalysis is the increase in the rate of a process by an "enzyme", a biological molecule. Most enzymes are proteins, and most such processes are chemical reactions. Within the enzyme, generally catalysis occurs at a localized site, calle ...
. Protein dynamics
In molecular biology, proteins are generally thought to adopt unique structures determined by their amino acid sequences. However, proteins are not strictly static objects, but rather populate ensembles of (sometimes similar) conformations. Tran ...
and conformational change
In biochemistry, a conformational change is a change in the shape of a macromolecule, often induced by environmental factors.
A macromolecule is usually flexible and dynamic. Its shape can change in response to changes in its environment or othe ...
s allow proteins to function as nanoscale biological machine
Molecular machines are a class of molecules typically described as an assembly of a discrete number of molecular components intended to produce mechanical movements in response to specific stimuli, mimicking macromolecular devices such as switch ...
s within cells, often in the form of multi-protein complexes. Examples include motor proteins
Motor proteins are a class of molecular motors that can move along the cytoskeleton of cells. They do this by converting chemical energy into mechanical work by the hydrolysis of ATP.
Cellular functions
Motor proteins are the driving force b ...
, such as myosin
Myosins () are a Protein family, family of motor proteins (though most often protein complexes) best known for their roles in muscle contraction and in a wide range of other motility processes in eukaryotes. They are adenosine triphosphate, ATP- ...
, which is responsible for muscle
Muscle is a soft tissue, one of the four basic types of animal tissue. There are three types of muscle tissue in vertebrates: skeletal muscle, cardiac muscle, and smooth muscle. Muscle tissue gives skeletal muscles the ability to muscle contra ...
contraction, kinesin
A kinesin is a protein complex belonging to a class of motor proteins found in eukaryotic cells. Kinesins move along microtubule (MT) filaments and are powered by the hydrolysis of adenosine triphosphate (ATP) (thus kinesins are ATPases, a typ ...
, which moves cargo inside cells away from the nucleus
Nucleus (: nuclei) is a Latin word for the seed inside a fruit. It most often refers to:
*Atomic nucleus, the very dense central region of an atom
*Cell nucleus, a central organelle of a eukaryotic cell, containing most of the cell's DNA
Nucleu ...
along microtubules
Microtubules are polymers of tubulin that form part of the cytoskeleton and provide structure and shape to eukaryotic cells. Microtubules can be as long as 50 micrometres, as wide as 23 to 27 nm and have an inner diameter between 11 an ...
, and dynein
Dyneins are a family of cytoskeletal motor proteins (though they are actually protein complexes) that move along microtubules in cells. They convert the chemical energy stored in ATP to mechanical work. Dynein transports various cellular cargo ...
, which moves cargo inside cells towards the nucleus and produces the axonemal beating of motile cilia
The cilium (: cilia; ; in Medieval Latin and in anatomy, ''cilium'') is a short hair-like membrane protrusion from many types of eukaryotic cell. (Cilia are absent in bacteria and archaea.) The cilium has the shape of a slender threadlike pr ...
and flagella
A flagellum (; : flagella) (Latin for 'whip' or 'scourge') is a hair-like appendage that protrudes from certain plant and animal sperm cells, from fungal spores ( zoospores), and from a wide range of microorganisms to provide motility. Many pr ...
. " effect, the otile ciliumis a nanomachine composed of perhaps over 600 proteins in molecular complexes, many of which also function independently as nanomachines...Flexible linker
In molecular biology, an intrinsically disordered protein (IDP) is a protein that lacks a fixed or ordered three-dimensional structure, typically in the absence of its macromolecular interaction partners, such as other proteins or RNA. IDPs rang ...
s allow the mobile protein domains connected by them to recruit their binding partners and induce long-range allostery
In the fields of biochemistry and pharmacology an allosteric regulator (or allosteric modulator) is a substance that binds to a site on an enzyme or Receptor (biochemistry), receptor distinct from the active site, resulting in a conformational ...
via protein domain dynamics. "
 Proteins are often thought of as relatively stable tertiary structures that experience conformational changes after being affected by interactions with other proteins or as a part of enzymatic activity. However, proteins may have varying degrees of stability, and some of the less stable variants are
Proteins are often thought of as relatively stable tertiary structures that experience conformational changes after being affected by interactions with other proteins or as a part of enzymatic activity. However, proteins may have varying degrees of stability, and some of the less stable variants are intrinsically disordered proteins
In molecular biology, an intrinsically disordered protein (IDP) is a protein that lacks a fixed or ordered protein tertiary structure, three-dimensional structure, typically in the absence of its macromolecular interaction partners, such as other ...
. These proteins exist and function in a relatively 'disordered' state lacking a stable tertiary structure
Protein tertiary structure is the three-dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains and the ...
. As a result, they are difficult to describe by a single fixed tertiary structure
Protein tertiary structure is the three-dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains and the ...
. Conformational ensembles
In protein chemistry, conformational ensembles, also known as structural ensembles, are models describing the structure of intrinsically unstructured proteins. Such proteins are flexible in nature and cannot be described by a single structur ...
have been devised as a way to provide a more accurate and 'dynamic' representation of the conformational state of intrinsically disordered proteins
In molecular biology, an intrinsically disordered protein (IDP) is a protein that lacks a fixed or ordered protein tertiary structure, three-dimensional structure, typically in the absence of its macromolecular interaction partners, such as other ...
.
Protein ensemble files are a representation of a protein that can be considered to have a flexible structure. Creating these files requires determining which of the various theoretically possible protein conformations actually exist. One approach is to apply computational algorithms to the protein data in order to try to determine the most likely set of conformations for an ensemble file. There are multiple methods for preparing data for thProtein Ensemble Database
that fall into two general methodologies – pool and molecular dynamics (MD) approaches (diagrammed in the figure). The pool based approach uses the protein's amino acid sequence to create a massive pool of random conformations. This pool is then subjected to more computational processing that creates a set of theoretical parameters for each conformation based on the structure. Conformational subsets from this pool whose average theoretical parameters closely match known experimental data for this protein are selected. The alternative molecular dynamics approach takes multiple random conformations at a time and subjects all of them to experimental data. Here the experimental data is serving as limitations to be placed on the conformations (e.g. known distances between atoms). Only conformations that manage to remain within the limits set by the experimental data are accepted. This approach often applies large amounts of experimental data to the conformations which is a very computationally demanding task. The conformational ensembles were generated for a number of highly dynamic and partially unfolded proteins, such as
Sic1
Sic1, a protein, is a stoichiometric inhibitor of Cdk1-Clb ( B-type cyclins) complexes in the budding yeast ''Saccharomyces cerevisiae''. Because B-type cyclin-Cdk1 complexes are the drivers of S-phase initiation, Sic1 prevents premature S-phase e ...
/ Cdc4, p15 PAF, MKK7
Dual specificity mitogen-activated protein kinase kinase 7, also known as MAP kinase kinase 7 or MKK7, is an enzyme that in humans is encoded by the ''MAP2K7'' gene. This protein is a member of the MKK, mitogen-activated protein kinase kinase fam ...
, Beta-synuclein and P27
Protein folding
As it is translated, polypeptides exit theribosome
Ribosomes () are molecular machine, macromolecular machines, found within all cell (biology), cells, that perform Translation (biology), biological protein synthesis (messenger RNA translation). Ribosomes link amino acids together in the order s ...
mostly as a random coil
In polymer chemistry, a random coil is a conformation of polymers where the monomer subunits are oriented randomly while still being bonded to adjacent units. It is not one specific shape, but a statistical distribution of shapes for all the cha ...
and folds into its native state
In biochemistry, the native state of a protein or nucleic acid is its properly Protein folding, folded and/or assembled form, which is operative and functional. The native state of a biomolecule may possess all four levels of biomolecular structu ...
. The final structure of the protein chain is generally assumed to be determined by its amino acid sequence (Anfinsen's dogma
Anfinsen's dogma, also known as the thermodynamic hypothesis, is a postulate in molecular biology. It states that, at least for a small globular protein in its standard physiological environment, the native structure is determined only by the pro ...
).
Protein stability
Thermodynamic stability of proteins represents the free energy difference between the folded and unfolded protein states. This free energy difference is very sensitive to temperature, hence a change in temperature may result in unfolding or denaturation.Protein denaturation
In biochemistry, denaturation is a process in which proteins or nucleic acids lose folded structure present in their native state due to various factors, including application of some external stress or compound, such as a strong acid or base ...
may result in loss of function, and loss of native state. The free energy of stabilization of soluble globular proteins typically does not exceed 50 kJ/mol. Taking into consideration the large number of hydrogen bonds that take place for the stabilization of secondary structures, and the stabilization of the inner core through hydrophobic interactions, the free energy of stabilization emerges as small difference between large numbers.
Protein structure determination

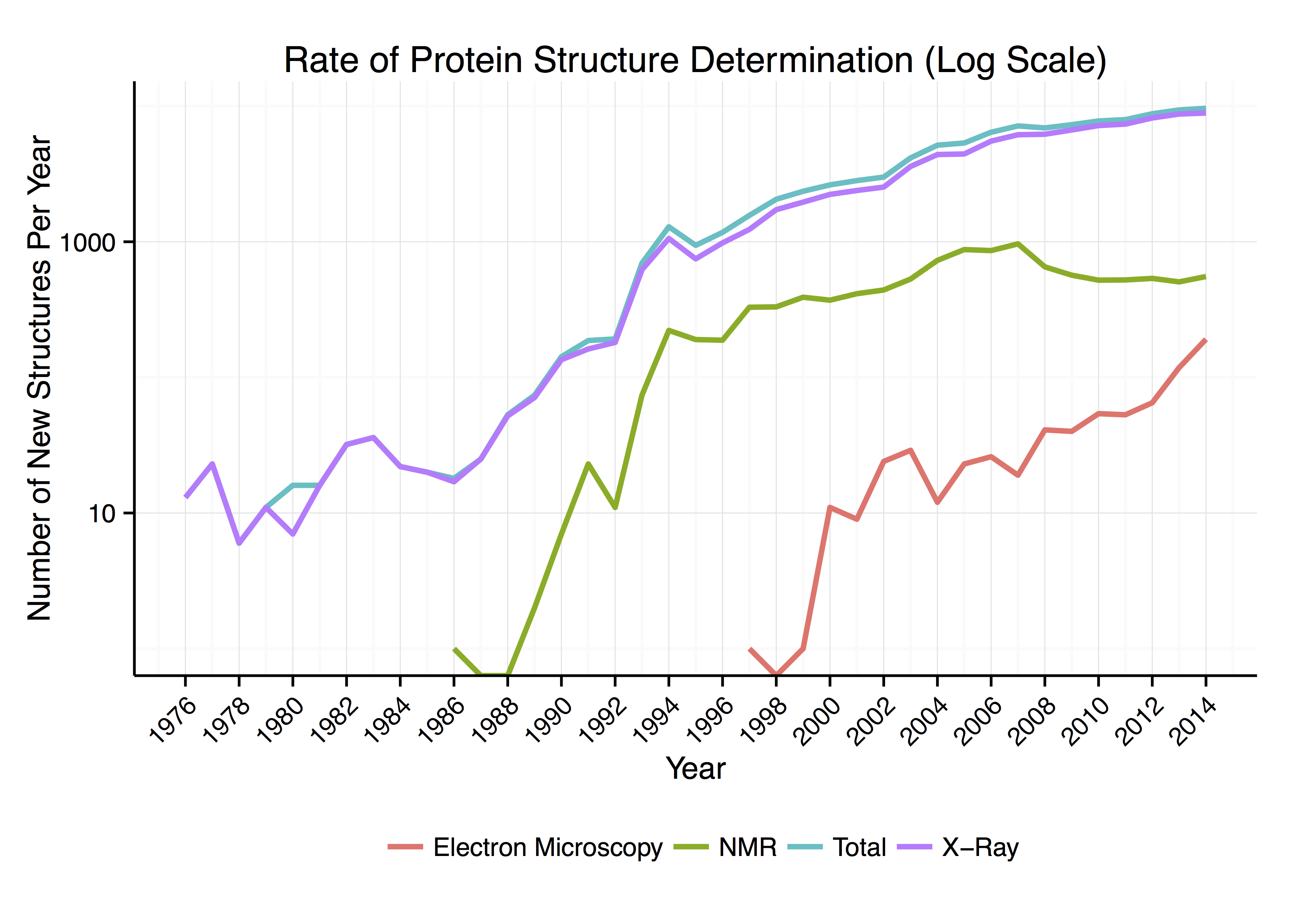
 Around 90% of the protein structures available in the
Around 90% of the protein structures available in the Protein Data Bank
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules such as proteins and nucleic acids, which is overseen by the Worldwide Protein Data Bank (wwPDB). This structural data is obtained a ...
have been determined by X-ray crystallography
X-ray crystallography is the experimental science of determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to Diffraction, diffract in specific directions. By measuring th ...
. This method allows one to measure the three-dimensional (3-D) density distribution of electron
The electron (, or in nuclear reactions) is a subatomic particle with a negative one elementary charge, elementary electric charge. It is a fundamental particle that comprises the ordinary matter that makes up the universe, along with up qua ...
s in the protein, in the crystallized state, and thereby infer
Inferences are steps in logical reasoning, moving from premises to logical consequences; etymologically, the word '' infer'' means to "carry forward". Inference is theoretically traditionally divided into deduction and induction, a distinctio ...
the 3-D coordinates of all the atom
Atoms are the basic particles of the chemical elements. An atom consists of a atomic nucleus, nucleus of protons and generally neutrons, surrounded by an electromagnetically bound swarm of electrons. The chemical elements are distinguished fr ...
s to be determined to a certain resolution. Roughly 7% of the known protein structures have been obtained by nuclear magnetic resonance
Nuclear magnetic resonance (NMR) is a physical phenomenon in which nuclei in a strong constant magnetic field are disturbed by a weak oscillating magnetic field (in the near field) and respond by producing an electromagnetic signal with a ...
(NMR) techniques. For larger protein complexes, cryo-electron microscopy
Cryogenic electron microscopy (cryo-EM) is a transmission electron microscopy technique applied to samples cooled to cryogenic temperatures. For biological specimens, the structure is preserved by embedding in an environment of vitreous ice. An ...
can determine protein structures. The resolution is typically lower than that of X-ray crystallography, or NMR, but the maximum resolution is steadily increasing. This technique is still a particularly valuable for very large protein complexes such as virus coat proteins and amyloid
Amyloids are aggregates of proteins characterised by a fibrillar morphology of typically 7–13 nm in diameter, a β-sheet secondary structure (known as cross-β) and ability to be stained by particular dyes, such as Congo red. In the human ...
fibers.
General secondary structure composition can be determined via circular dichroism
Circular dichroism (CD) is dichroism involving circular polarization, circularly polarized light, i.e., the differential Absorption (electromagnetic radiation), absorption of left- and right-handed light. Left-hand circular (LHC) and right-hand ci ...
. Vibrational spectroscopy
Infrared spectroscopy (IR spectroscopy or vibrational spectroscopy) is the measurement of the interaction of infrared radiation with matter by absorption, emission, or reflection. It is used to study and identify chemical substances or functio ...
can also be used to characterize the conformation of peptides, polypeptides, and proteins. Two-dimensional infrared spectroscopy has become a valuable method to investigate the structures of flexible peptides and proteins that cannot be studied with other methods. A more qualitative picture of protein structure is often obtained by proteolysis
Proteolysis is the breakdown of proteins into smaller polypeptides or amino acids. Protein degradation is a major regulatory mechanism of gene expression and contributes substantially to shaping mammalian proteomes. Uncatalysed, the hydrolysis o ...
, which is also useful to screen for more crystallizable protein samples. Novel implementations of this approach, including fast parallel proteolysis (FASTpp), can probe the structured fraction and its stability without the need for purification. Once a protein's structure has been experimentally determined, further detailed studies can be done computationally, using molecular dynamic simulations of that structure.
Protein structure databases
Aprotein structure database
In biology, a protein structure database is a database that is modeled around the various experimentally determined protein structures. The aim of most protein structure databases is to organize and annotate the protein structures, providing the ...
is a database that is modeled around the various experimentally determined protein structures. The aim of most protein structure databases is to organize and annotate the protein structures, providing the biological community access to the experimental data in a useful way. Data included in protein structure databases often includes 3D coordinates as well as experimental information, such as unit cell dimensions and angles for x-ray crystallography
X-ray crystallography is the experimental science of determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to Diffraction, diffract in specific directions. By measuring th ...
determined structures. Though most instances, in this case either proteins or a specific structure determinations of a protein, also contain sequence information and some databases even provide means for performing sequence based queries, the primary attribute of a structure database is structural information, whereas sequence database
In the field of bioinformatics, a sequence database is a type of biological database that is composed of a large collection of computerized ("Digital data, digital") nucleic acid sequences, protein sequences, or other polymer sequences stored on a ...
s focus on sequence information, and contain no structural information for the majority of entries. Protein structure databases are critical for many efforts in computational biology
Computational biology refers to the use of techniques in computer science, data analysis, mathematical modeling and Computer simulation, computational simulations to understand biological systems and relationships. An intersection of computer sci ...
such as structure based drug design, both in developing the computational methods used and in providing a large experimental dataset used by some methods to provide insights about the function of a protein.
Structural classifications of proteins
Protein structures can be grouped based on their structural similarity, topological class or a commonevolution
Evolution is the change in the heritable Phenotypic trait, characteristics of biological populations over successive generations. It occurs when evolutionary processes such as natural selection and genetic drift act on genetic variation, re ...
ary origin. The Structural Classification of Proteins database
The Structural Classification of Proteins (SCOP) database is a largely manual classification of protein structural domains based on similarities of their structures and amino acid sequences. A motivation for this classification is to determine t ...
and CATH Cath may refer to:
__NOTOC__ People
* Cath Bishop (born 1971), British former rower and 2003 world champion
* Cath Carroll (born 1960), British musician and music journalist
* Cath Coffey (), one of the earliest members of British rap band Stereo ...
database provide two different structural classifications of proteins. When the structural similarity is large the two proteins have possibly diverged from a common ancestor, and shared structure between proteins is considered evidence of homology. Structure similarity can then be used to group proteins together into protein superfamilies
A protein superfamily is the largest grouping (clade) of proteins for which common ancestry can be inferred (see homology (biology), homology). Usually this common ancestry is inferred from structural alignment and mechanistic similarity, even if n ...
. If shared structure is significant but the fraction shared is small, the fragment shared may be the consequence of a more dramatic evolutionary event such as horizontal gene transfer
Horizontal gene transfer (HGT) or lateral gene transfer (LGT) is the movement of genetic material between organisms other than by the ("vertical") transmission of DNA from parent to offspring (reproduction). HGT is an important factor in the e ...
, and joining proteins sharing these fragments into protein superfamilies is no longer justified. Topology of a protein can be used to classify proteins as well. Knot theory
In topology, knot theory is the study of knot (mathematics), mathematical knots. While inspired by knots which appear in daily life, such as those in shoelaces and rope, a mathematical knot differs in that the ends are joined so it cannot be und ...
and circuit topology
The circuit topology of a folded linear polymer refers to the arrangement of its intra-molecular contacts. Examples of linear polymers with intra-molecular contacts are nucleic acids and proteins. Proteins fold via the formation of contacts of v ...
are two topology frameworks developed for classification of protein folds based on chain crossing and intrachain contacts respectively.
Computational prediction of protein structure
The generation of aprotein sequence
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthe ...
is much easier than the determination of a protein structure. However, the structure of a protein gives much more insight in the function of the protein than its sequence. Therefore, a number of methods for the computational prediction of protein structure from its sequence have been developed. ''Ab initio'' prediction methods use just the sequence of the protein. Threading and homology modeling
Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the "''target''" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous pr ...
methods can build a 3-D model for a protein of unknown structure from experimental structures of evolutionarily-related proteins, called a protein family
A protein family is a group of evolutionarily related proteins. In many cases, a protein family has a corresponding gene family, in which each gene encodes a corresponding protein with a 1:1 relationship. The term "protein family" should not be ...
.
See also
*Biomolecular structure
Biomolecular structure is the intricate folded, three-dimensional shape that is formed by a molecule of protein, DNA, or RNA, and that is important to its function. The structure of these molecules may be considered at any of several length sca ...
* Gene structure
Gene structure is the organisation of specialised sequence elements within a gene. Genes contain most of the information necessary for living cells to survive and reproduce. In most organisms, genes are made of DNA, where the particular DNA sequen ...
* Nucleic acid structure
Nucleic acid structure refers to the structure of nucleic acids such as DNA and RNA. Chemically speaking, DNA and RNA are very similar. Nucleic acid structure is often divided into four different levels: primary, secondary, tertiary, and quaterna ...
* PCRPi-DB
* Ribbon diagram
Ribbon diagrams, also known as Richardson diagrams, are three-dimensional space, 3D schematic representations of protein structure and are one of the most common methods of protein depiction used today. The ribbon depicts the general course and o ...
3D schematic representation of proteins
References
Further reading
50 Years of Protein Structure Determination Timeline - HTML Version - National Institute of General Medical Sciences
at
NIH
The National Institutes of Health (NIH) is the primary agency of the United States government responsible for biomedical and public health research. It was founded in 1887 and is part of the United States Department of Health and Human Service ...
External links
* {{DEFAULTSORT:Protein StructureProtein Structure
drugdesign.org
Method_for_the_Characterization_of_the_Three-Dimensional_Structure_of_Proteins_Employing_Mass_Spectrometric_Analysis_and_Experimental-Computational_Feedback_Modeling
A_Method_for_the_Determination_of_the_Conformation_(Topology)_of_Proteins_Employing_Experimental-Computational_Feedback_Modeling