|
Loop Extrusion
Loop extrusion is a major mechanism of Nuclear organization. It is a dynamic process in which structural maintenance of chromosomes (SMC) protein complexes progressively grow loops of DNA or chromatin. In this process, SMC complexes, such as condensin or cohesin, bind to DNA/chromatin, use ATP-driven motor activity to reel in DNA, and as a result, extrude the collected DNA as a loop. Background The organization of DNA presents a remarkable biological challenge: human DNA can reach 2 meters and is packed into the nucleus with the diameter of 5-20 μm. At the same time, the critical cell processes involve complex processes on highly compacted DNA, such as transcription, replication, recombination, DNA repair, and cell division. Loop extrusion is a key mechanism that organizes DNA into loops, enabling its efficient compaction and functional organization. For instance, ''in vitro'' experiments show that cohesin can compact DNA by 80%, while condensin achieves a remarkable 10 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important roles in reinforcing the DNA during cell division, preventing DNA repair#DNA damage, DNA damage, and regulating gene expression and DNA replication. During mitosis and meiosis, chromatin facilitates proper segregation of the chromosomes in anaphase; the characteristic shapes of chromosomes visible during this stage are the result of DNA being coiled into highly condensed chromatin. The primary protein components of chromatin are histones. An octamer of two sets of four histone cores (Histone H2A, Histone H2B, Histone H3, and Histone H4) bind to DNA and function as "anchors" around which the strands are wound.Maeshima, K., Ide, S., & Babokhov, M. (2019). Dynamic chromatin organization without the 30 nm fiber. ''Current opinion in cell biolog ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Loop Extrusion
Loop extrusion is a major mechanism of Nuclear organization. It is a dynamic process in which structural maintenance of chromosomes (SMC) protein complexes progressively grow loops of DNA or chromatin. In this process, SMC complexes, such as condensin or cohesin, bind to DNA/chromatin, use ATP-driven motor activity to reel in DNA, and as a result, extrude the collected DNA as a loop. Background The organization of DNA presents a remarkable biological challenge: human DNA can reach 2 meters and is packed into the nucleus with the diameter of 5-20 μm. At the same time, the critical cell processes involve complex processes on highly compacted DNA, such as transcription, replication, recombination, DNA repair, and cell division. Loop extrusion is a key mechanism that organizes DNA into loops, enabling its efficient compaction and functional organization. For instance, ''in vitro'' experiments show that cohesin can compact DNA by 80%, while condensin achieves a remarkable 10 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CTCF
Transcriptional repressor CTCF also known as 11-zinc finger protein or CCCTC-binding factor is a transcription factor that in humans is encoded by the ''CTCF'' gene. CTCF is involved in many cellular processes, including transcriptional regulation, insulator activity, V(D)J recombination and regulation of chromatin architecture. Discovery CCCTC-Binding factor or CTCF was initially discovered as a negative regulator of the chicken c-myc gene. This protein was found to be binding to three regularly spaced repeats of the core sequence CCCTC and thus was named CCCTC binding factor. Function The primary role of CTCF is thought to be in regulating the 3D structure of chromatin. CTCF binds together strands of DNA, thus forming chromatin loops, and anchors DNA to cellular structures like the nuclear lamina. It also defines the boundaries between active and heterochromatic DNA. Since the 3D structure of DNA influences the regulation of genes, CTCF's activity influences the expre ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
V(D)J Recombination
V(D)J recombination (variable–diversity–joining rearrangement) is the mechanism of somatic recombination that occurs only in developing lymphocytes during the early stages of T and B cell maturation. It results in the highly diverse repertoire of Antibody, antibodies/immunoglobulins and T cell receptors (TCRs) found in B cells and T cells, respectively. The process is a defining feature of the adaptive immune system. V(D)J recombination in mammals occurs in the primary lymphoid organs (bone marrow for B cells and thymus for T cells) and in a nearly random fashion rearranges variable (V), joining (J), and in some cases, diversity (D) gene segments. The process ultimately results in novel amino acid sequences in the antigen-binding regions of immunoglobulins and TCRs that allow for the recognition of antigens from nearly all pathogens including bacteria, viruses, parasites, and Helminths, worms as well as "altered self cells" as seen in cancer. The recognition can also be Allergy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cell Division
Cell division is the process by which a parent cell (biology), cell divides into two daughter cells. Cell division usually occurs as part of a larger cell cycle in which the cell grows and replicates its chromosome(s) before dividing. In eukaryotes, there are two distinct types of cell division: a vegetative division (mitosis), producing daughter cells genetically identical to the parent cell, and a cell division that produces Haploidisation, haploid gametes for sexual reproduction (meiosis), reducing the number of chromosomes from two of each type in the diploid parent cell to one of each type in the daughter cells. Mitosis is a part of the cell cycle, in which, replicated chromosomes are separated into two new Cell nucleus, nuclei. Cell division gives rise to genetically identical cells in which the total number of chromosomes is maintained. In general, mitosis (division of the nucleus) is preceded by the S stage of interphase (during which the DNA replication occurs) and is f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mitosis
Mitosis () is a part of the cell cycle in eukaryote, eukaryotic cells in which replicated chromosomes are separated into two new Cell nucleus, nuclei. Cell division by mitosis is an equational division which gives rise to genetically identical cells in which the total number of chromosomes is maintained. Mitosis is preceded by the S phase of interphase (during which DNA replication occurs) and is followed by telophase and cytokinesis, which divide the cytoplasm, organelles, and cell membrane of one cell into two new cell (biology), cells containing roughly equal shares of these cellular components. The different stages of mitosis altogether define the mitotic phase (M phase) of a cell cycle—the cell division, division of the mother cell into two daughter cells genetically identical to each other. The process of mitosis is divided into stages corresponding to the completion of one set of activities and the start of the next. These stages are preprophase (specific to plant ce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
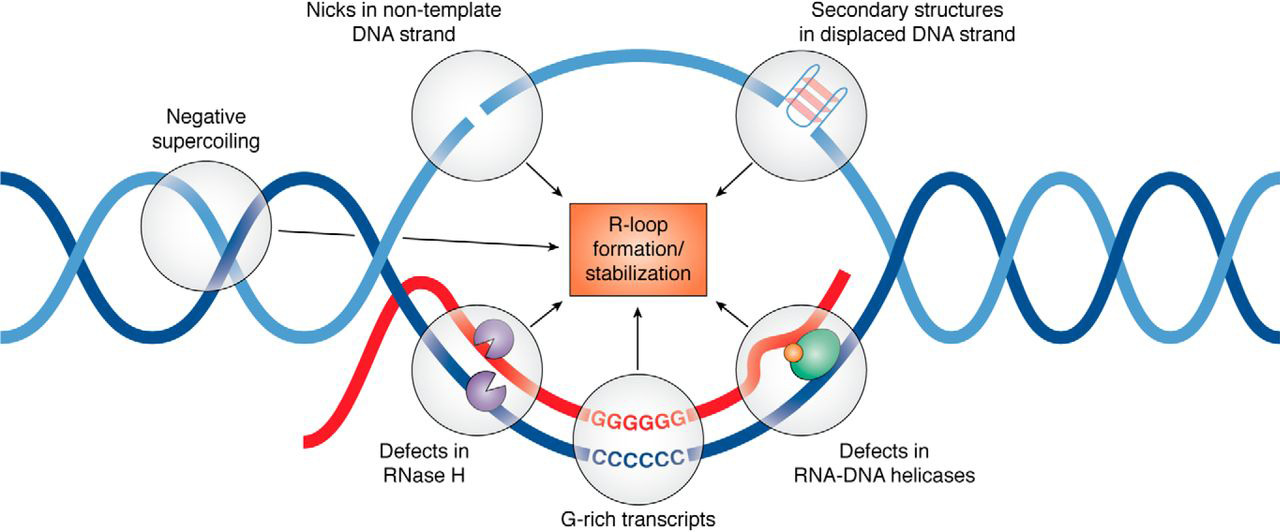
R-loop
An R-loop is a three-stranded nucleic acid structure, composed of a DNA:RNA hybrid and the associated non-template single-stranded DNA. R-loops may be formed in a variety of circumstances and may be tolerated or cleared by cellular components. The term "R-loop" was given to reflect the similarity of these structures to D-loops; the "R" in this case represents the involvement of an RNA moiety (chemistry), moiety. In the laboratory, R-loops can be created by transcription of DNA sequences (for example those that have a high GC content) that favor annealing of the RNA behind the progressing RNA polymerase. At least 100bp of DNA:RNA hybrid is required to form a stable R-loop structure. R-loops may also be created by the Nucleic acid hybridization#Hybridization, hybridization of mature mRNA with double-stranded DNA under conditions favoring the formation of a DNA-RNA hybrid; in this case, the intron regions (which have been Splicing (genetics), spliced out of the mRNA) form single-stra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Helicase
Helicases are a class of enzymes that are vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic double helix, separating the two hybridized nucleic acid strands (hence '' helic- + -ase''), via the energy gained from ATP hydrolysis. There are many helicases, representing the great variety of processes in which strand separation must be catalyzed. Approximately 1% of eukaryotic genes code for helicases. The human genome codes for 95 non-redundant helicases: 64 RNA helicases and 31 DNA helicases. Many cellular processes, such as DNA replication, transcription, translation, recombination, DNA repair and ribosome biogenesis involve the separation of nucleic acid strands that necessitates the use of helicases. Some specialized helicases are also involved in sensing viral nucleic acids during infection and fulfill an immunological function. Genetic mutations that affect helicase ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase
In biochemistry, a polymerase is an enzyme (Enzyme Commission number, EC 2.7.7.6/7/19/48/49) that synthesizes long chains of polymers or nucleic acids. DNA polymerase and RNA polymerase are used to assemble DNA and RNA molecules, respectively, by copying a DNA template strand using Base pair, base-pairing interactions or RNA by half ladder replication. A DNA polymerase from the thermophile, thermophilic bacterium, ''Thermus aquaticus'' (''Taq'') (Protein Data Bank, PDB]1BGX EC 2.7.7.7) is used in the polymerase chain reaction, an important technique of molecular biology. A polymerase may be template-dependent or template-independent. Polynucleotide adenylyltransferase, Poly-A-polymerase is an example of template independent polymerase. Terminal deoxynucleotidyl transferase also known to have template independent and template dependent activities. By function *DNA polymerase (DNA-directed DNA polymerase, DdDP) **Family A: DNA polymerase I; Pol POLG, γ, POLQ, θ, DNA polymer ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Topologically Associating Domain
A topologically associating domain (TAD) is a self-interacting genomic region, meaning that DNA sequences within a TAD physically interact with each other more frequently than with sequences outside the TAD. The average size of a topologically associating domain (TAD) is 1000 kb in humans, 880 kb in mouse cells, and 140 kb in fruit flies. Boundaries at both side of these domains are conserved between different mammalian cell types and even across species and are highly enriched with CTCF, CCCTC-binding factor (CTCF) and cohesin. In addition, some types of genes (such as transfer RNA genes and housekeeping genes) appear near TAD boundaries more often than would be expected by chance. The functions of TADs are not fully understood and are still a matter of debate. Most of the studies indicate TADs regulate gene expression by limiting the Enhancer (genetics), enhancer-Promoter (genetics), promoter interaction to each TAD; however, a recent study uncouples TAD organization and gene exp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
STAG2
Cohesin subunit SA-2 (SA2) is a protein that in humans is encoded by the ''STAG2'' gene. SA2 is a subunit of the Cohesin complex which mediates sister chromatid cohesion, homologous recombination and DNA looping. In somatic cells cohesin is formed of SMC3, SMC1, RAD21 and either SA1 or SA2 whereas in meiosis, cohesin is formed of SMC3, SMC1B, REC8 and SA3. ''STAG2'' is frequently mutated in a range of cancers and several other disorders. Function SA2 is part of the cohesin complex, which is a structure that holds the sister chromatids together after DNA replication. STAG2 has been shown to interact with STAG1. Cohesion folds by DNA loop extrusion and this cohesion consists of SMC1, SMC3, RAD21, and either STAG1 or STAG2. SA2 interacts with a ring-like structure composed of SMC1A, SMC3, and RAD21, to form the core of the cohesin complex. The ring-like structure binds chromosomes together until degradation of the cohesin complex is finished during cell division. This ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ESCO1
Establishment of sister chromatid cohesion N-acetyltransferase 1 is a protein that in humans is encoded by the ESCO1 gene. Function ESCO1 belongs to a conserved family of acetyltransferases involved in sister chromatid A sister chromatid refers to the identical copies ( chromatids) formed by the DNA replication of a chromosome, with both copies joined together by a common centromere. In other words, a sister chromatid may also be said to be 'one-half' of the du ... cohesion. See also * Lysine N-acetyltransferase References External links PDBe-KB provides an overview of all the structure information available in the PDB for Human N-acetyltransferase ESCO1 {{gene-18-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |