sucrase-isomaltase on:
[Wikipedia]
[Google]
[Amazon]
Oligo-1,6-glucosidase (EC 3.2.1.10, sucrase-isomaltase, SI; systematic name oligosaccharide 6-α-glucohydrolase) is a
 Sucrase-isomaltase consists of two enzymatic subunits: sucrase and
Sucrase-isomaltase consists of two enzymatic subunits: sucrase and
Structure and evolution of the mammalian maltase-glucoamylase and sucrase-isomaltase
* {{Portal bar, Biology, border=no EC 3.2.1
glucosidase
Glucosidases are the glycoside hydrolase enzymes categorized under the EC number 3.2.1.
Function
Alpha-glucosidases are enzymes involved in breaking down complex carbohydrates such as starch and glycogen into their monomers.
They catalyze ...
enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products ...
located on the brush border of the small intestine, which catalyses the following reaction:
:Hydrolysis of (1→6)-α-D-glucosidic linkages in some oligosaccharides produced from starch and glycogen by (α-amylase), and in isomaltose
It is a dual-function enzyme with two GH31 domains, one serving as the isomaltase
Isomaltase () is an enzyme that breaks the bonds linking saccharides, which cannot be broken by amylase or maltase. It digests polysaccharides at the alpha 1-6 linkages. Its substrate, alpha-limit dextrin, is a product of amylopectin digestion t ...
, the other as a sucrose alpha-glucosidase. It has preferential expression in the apical membranes of enterocytes
Enterocytes, or intestinal absorptive cells, are simple columnar epithelial cells which line the inner surface of the small and large intestines. A glycocalyx surface coat contains digestive enzymes. Microvilli on the apical surface increase it ...
. The enzyme’s purpose is to digest dietary carbohydrates such as starch, sucrose and isomaltose
Isomaltose is a disaccharide similar to maltose, but with a α-(1-6)-linkage instead of the α-(1-4)-linkage. Both of the sugars are dimers of glucose, which is a pyranose sugar. Isomaltose is a reducing sugar. Isomaltose is produced when high ...
. By further processing the broken-down products, energy in the form of ATP can be generated.Berg, J. M. et al. ''Biochemistry'', 7th Ed. W.H. Freeman and Company: New York, 2012.
Structure
 Sucrase-isomaltase consists of two enzymatic subunits: sucrase and
Sucrase-isomaltase consists of two enzymatic subunits: sucrase and isomaltase
Isomaltase () is an enzyme that breaks the bonds linking saccharides, which cannot be broken by amylase or maltase. It digests polysaccharides at the alpha 1-6 linkages. Its substrate, alpha-limit dextrin, is a product of amylopectin digestion t ...
. The subunits originate from a polypeptide precursor, pro-SI. By heterodimerizing the two subunits, the sucrase-isomaltase complex is formed. The enzyme is anchored in the intestinal brush border membrane by a hydrophobic segment located near the N-terminal of the isomaltase subunit. Before the enzyme is anchored to the membrane, pro-SI is mannose-rich and glycosylated; it moves from the ER to the Golgi, where it becomes a protein complex that is N- and O- glycosylated. The O-linked glycosylation is necessary to target the protein to the apical membrane. In addition, there is a segment that is both O-linked glycosylated and Ser/Thr-rich. A similarly-arranged enzyme is the maltase-glucoamylase
Maltase-glucoamylase, intestinal is an enzyme that in humans is encoded by the ''MGAM'' gene.
Maltase-glucoamylase is an alpha-glucosidase digestive enzyme. It consists of two subunits with differing substrate specificity. Recombinant enzyme stu ...
, also a member of GH31.
Sucrase-isomaltase is composed of duplicated catalytic domains, N- and C-terminal. Each domain displays overlapping specificities. Scientists have discovered the crystal structure for N-terminal human sucrase-isomaltase (ntSI) in apo form to 3.2 Å and in complex with the inhibitor kotalanol to 2.15 Å resolution. Sucrase-isomaltase’s mechanism results in a net retention of configuration at the anomeric center.
The crystal structure shows that sucrase-isomaltase exists as a monomer
In chemistry, a monomer ( ; '' mono-'', "one" + ''-mer'', "part") is a molecule that can react together with other monomer molecules to form a larger polymer chain or three-dimensional network in a process called polymerization.
Classification
...
. The researchers claim that the observance of SI dimers is dependent on experimental conditions. ntSI’s four monomers, A, B, C, and D are included in the crystal asymmetric unit and have identical active sites. The active site is composed of a shallow-substrate binding pocket including -1 and +1 subsites. The non-reducing end of substrates binds to the pocket. While the non-reducing sugar ring has interactions with the buried -1 subsite, the reducing ring has interactions with the surface exposed +1 subsite.
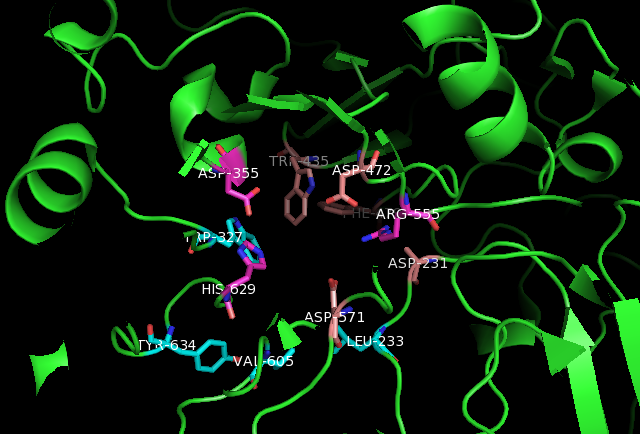
The interactions between the active site of sucrase-isomaltase and the following compounds have been identified:
* Man2GlcNAc2 glycan: Within the active site, Man2GlcNAc2 hydrogen bonds with hydroxyl
In chemistry, a hydroxy or hydroxyl group is a functional group with the chemical formula and composed of one oxygen atom covalently bonded to one hydrogen atom. In organic chemistry, alcohols and carboxylic acids contain one or more hydro ...
side chains of Asp231 and Asp571. Furthermore, hydrophobic interactions with Leu233, Trp327, Trp435, Phe479, Val605, and Tyr634 provide additional stabilization for Man2GlcNAc2.
* Kotalonal, the inhibitor: It interacts with the catalytic nucleophile Asp472 and acid base catalyst Asp571. In addition, ntSI residues His629, Asp355, Arg555, Asp231, Trp435, and Phe479 bind to the substrate.
Currently, there are no crystal structures of ntSI in complex with an α-1,6-linked substrate or inhibitor analogue. In order to predict isomaltose binding in sucrase-isomaltase structure, a model was produced by hand. Within the -1 subsite, isomaltose’s non-reducing glucose ring was aligned to that of acarbose.
Not only has the structure of human sucrase-isomaltase been studied, but also sucrase-isomaltase’s structure in sea lions and pigs have also been analyzed.
Disease relevance
A deficiency is responsible for sucrose intolerance. Congenital sucrase-isomaltase deficiency (CSID), also called genetic sucrase-isomaltase deficiency (GSID), and sucrose intolerance, is a genetic, intestinal disorder that is caused by a reduction or absence of sucrase andisomaltase
Isomaltase () is an enzyme that breaks the bonds linking saccharides, which cannot be broken by amylase or maltase. It digests polysaccharides at the alpha 1-6 linkages. Its substrate, alpha-limit dextrin, is a product of amylopectin digestion t ...
Explanations for GSID include:
* Mutations C1229Y and F1745C, which are present in the sucrase domain of SI, block SI path to anchor in the cell’s aprical membrane but does not impact protein folding or isomaltase activity.
* Substitution of a cysteine by an arginine at amino acid residue 635 in the isomaltase subunit of SI was present in the cDNA encoding for a patient with CSID. SIC635R had an altered folding pattern, which influenced the sorting profile and increased the turnover rate.
* A factor that can be attributed to congenital sucrase-isomaltase deficiency is the retention of SI in the cis-Golgi. This inability to transport is a result of a glutamine to proline substitution at amino acid residue 1098 of the sucrase subunit.
Furthermore, a relationship between mutations in sucrase-isomaltase and chronic lymphocytic leukemia (CLL) has been identified. These mutations cause a loss of enzyme function by blocking the biosynthesis of SI at the cell surface.
See also
* Sucrase *Isomaltase
Isomaltase () is an enzyme that breaks the bonds linking saccharides, which cannot be broken by amylase or maltase. It digests polysaccharides at the alpha 1-6 linkages. Its substrate, alpha-limit dextrin, is a product of amylopectin digestion t ...
* Brush border
A brush border (striated border or brush border membrane) is the microvilli-covered surface of simple cuboidal and simple columnar epithelium found in different parts of the body. Microvilli are approximately 100 nanometers in diameter and their ...
References
External links
Structure and evolution of the mammalian maltase-glucoamylase and sucrase-isomaltase
* {{Portal bar, Biology, border=no EC 3.2.1