Viral Replicase on:
[Wikipedia]
[Google]
[Amazon]
RNA-dependent RNA polymerase (RdRp) or RNA replicase is an enzyme that catalyzes the
 Viral/prokaryotic RNA-directed RNA polymerases, along with many single-subunit DNA-directed polymerases, employ a fold whose organization has been linked to the shape of a right hand with three subdomains termed fingers, palm, and thumb. Only the palm subdomain, composed of a four-stranded antiparallel
Viral/prokaryotic RNA-directed RNA polymerases, along with many single-subunit DNA-directed polymerases, employ a fold whose organization has been linked to the shape of a right hand with three subdomains termed fingers, palm, and thumb. Only the palm subdomain, composed of a four-stranded antiparallel
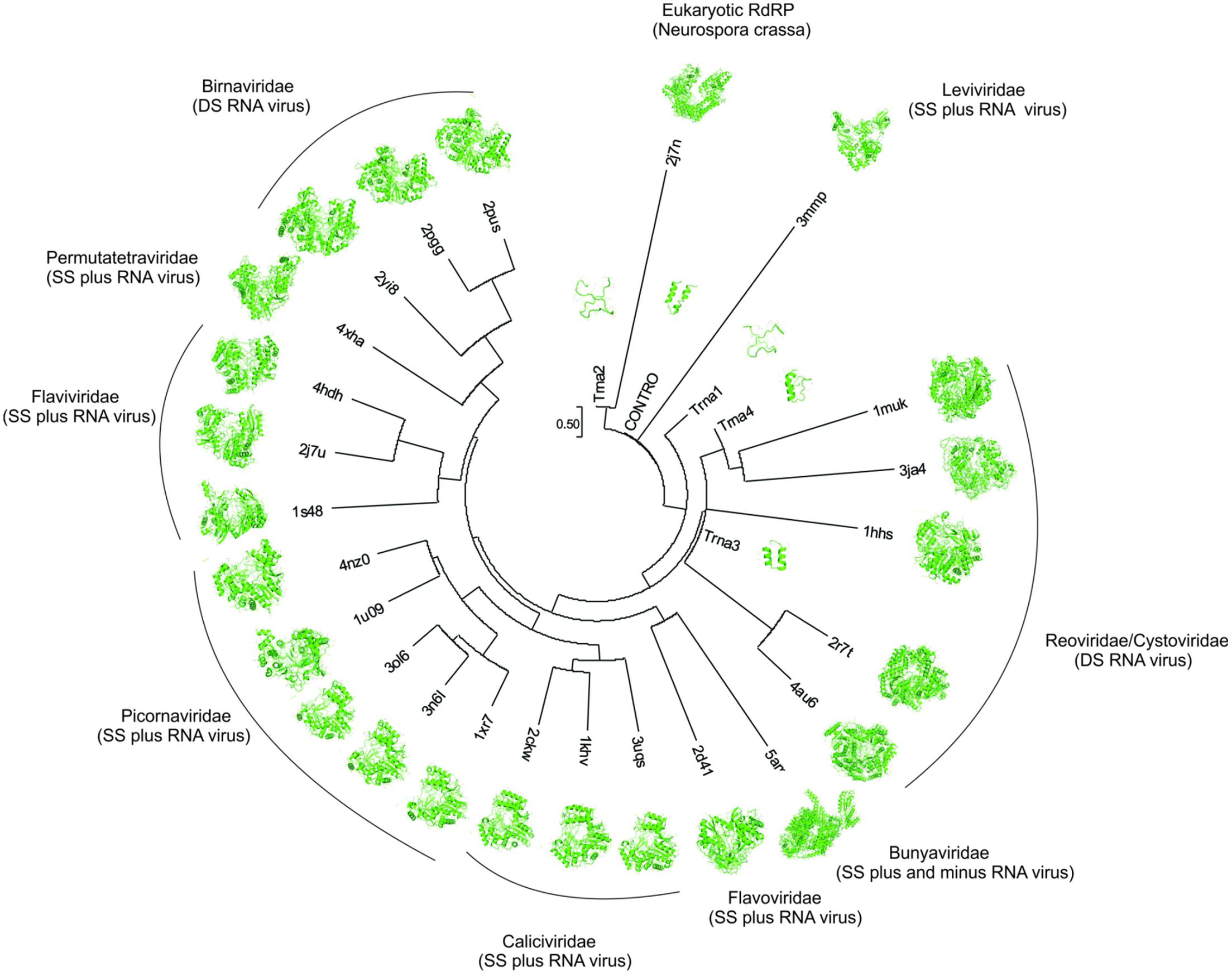 There are 4 superfamilies of viruses that cover all RNA-containing viruses with no DNA stage:
* Viruses containing positive-strand RNA or double-strand RNA, except
There are 4 superfamilies of viruses that cover all RNA-containing viruses with no DNA stage:
* Viruses containing positive-strand RNA or double-strand RNA, except
replication
Replication may refer to:
Science
* Replication (scientific method), one of the main principles of the scientific method, a.k.a. reproducibility
** Replication (statistics), the repetition of a test or complete experiment
** Replication crisi ...
of RNA
Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and deoxyribonucleic acid ( DNA) are nucleic acids. Along with lipids, proteins, and carbohydra ...
from an RNA template. Specifically, it catalyzes synthesis of the RNA strand complementary
A complement is something that completes something else.
Complement may refer specifically to:
The arts
* Complement (music), an interval that, when added to another, spans an octave
** Aggregate complementation, the separation of pitch-class ...
to a given RNA template. This is in contrast to typical DNA-dependent RNA polymerases, which all organisms use to catalyze the transcription of RNA from a DNA template.
RdRp is an essential protein encoded in the genomes of most RNA-containing viruses with no DNA stage including SARS-CoV-2. Some eukaryotes also contain RdRps, which are involved in RNA interference and differ structurally from viral RdRps.
History
Viral RdRps were discovered in the early 1960s from studies on mengovirus and polio virus when it was observed that these viruses were not sensitive toactinomycin D
Dactinomycin, also known as actinomycin D, is a chemotherapy medication used to treat a number of types of cancer. This includes Wilms tumor, rhabdomyosarcoma, Ewing's sarcoma, trophoblastic neoplasm, testicular cancer, and certain types of ovari ...
, a drug that inhibits cellular DNA-directed RNA synthesis. This lack of sensitivity suggested that there is a virus-specific enzyme that could copy RNA from an RNA template and not from a DNA template.
Distribution
RdRps are highly conserved throughout viruses and are even related to telomerase, though the reason for this is an ongoing question as of 2009. The similarity has led to speculation that viral RdRps are ancestral to human telomerase. The most famous example of RdRp is that of the polio virus. The viral genome is composed of RNA, which enters the cell through receptor-mediatedendocytosis
Endocytosis is a cellular process in which substances are brought into the cell. The material to be internalized is surrounded by an area of cell membrane, which then buds off inside the cell to form a vesicle containing the ingested material. E ...
. From there, the RNA is able to act as a template for complementary RNA synthesis, immediately. The complementary strand is then, itself, able to act as a template for the production of new viral genomes that are further packaged and released from the cell ready to infect more host cells. The advantage of this method of replication is that there is no DNA stage; replication is quick and easy. The disadvantage is that there is no 'back-up' DNA copy.
Many RdRps are associated tightly with membranes and are, therefore, difficult to study. The best-known RdRps are polioviral 3Dpol, vesicular stomatitis virus L, and hepatitis C virus NS5B protein.
Many eukaryote
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacte ...
s also have RdRps and these are involved in RNA interference: these amplify microRNAs and small temporal RNAs and produce double-stranded RNA using small interfering RNAs as primers. In fact these same RdRps that are used in the defense mechanisms can be usurped by RNA viruses for their benefit. Their evolutionary history has been reviewed.
Replication process
RdRp differs fromRNA polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template.
Using the enzyme helicase, RNAP locally opens the ...
as it works to catalyze the synthesis of an RNA strand complementary to a given RNA template, rather than using a DNA template. The RNA replication process is a four-step mechanism, as described.
# Nucleoside triphosphate (NTP) binding – initially, the RdRp presents with a vacant active site in which an NTP binds, complementary to the corresponding nucleotide on the template strand. Correct NTP binding causes the RdRp to undergo a conformational change.
# Active site closure – the conformational change, initiated by the correct NTP binding, results in the restriction of active site access and produces a catalytically competent state.
# Phosphodiester bond formation – two Mg2+ ions are present in the catalytically active state and arrange themselves in such a way around the newly synthesized RNA chain that the substrate NTP is able to undergo a phosphatidyl transfer and form a phosphodiester bond with the newly synthesized chain. Without the use of these Mg2+ ions, the active site is no longer catalytically stable and the RdRp complex changes to an open conformation.
# Translocation – once the active site is open, the RNA template strand is able to move by one position through the RdRp protein complex and continue chain elongation by binding a new NTP, unless otherwise specified by the template.
RNA synthesis can be performed by means of a primer
Primer may refer to:
Arts, entertainment, and media Films
* ''Primer'' (film), a 2004 feature film written and directed by Shane Carruth
* ''Primer'' (video), a documentary about the funk band Living Colour
Literature
* Primer (textbook), a t ...
-independent (''de novo'') or a primer-dependent mechanism that utilizes a viral protein genome-linked (VPg) primer. The ''de novo'' initiation consists in the addition of a nucleoside triphosphate (NTP) to the 3'-OH of the first initiating NTP. During the following so-called elongation phase, this nucleotidyl transfer reaction is repeated with subsequent NTPs to generate the complementary RNA product. Termination of the nascent RNA chain produced by RdRp is not completely known, however, it has been shown that RdRp termination is sequence-independent.
One major drawback of RNA-dependent RNA polymerase replication is the immense error rate during transcription. RdRps are known to have a lack of fidelity on the order of 104 nucleotides, which is thought to be a direct result of its insufficient proofreading abilities. This high rate of variation is favored in viral genomes as it allows for the pathogen to overcome defenses developed by hosts trying to avoid infection allowing for evolutionary growth.
Structure
 Viral/prokaryotic RNA-directed RNA polymerases, along with many single-subunit DNA-directed polymerases, employ a fold whose organization has been linked to the shape of a right hand with three subdomains termed fingers, palm, and thumb. Only the palm subdomain, composed of a four-stranded antiparallel
Viral/prokaryotic RNA-directed RNA polymerases, along with many single-subunit DNA-directed polymerases, employ a fold whose organization has been linked to the shape of a right hand with three subdomains termed fingers, palm, and thumb. Only the palm subdomain, composed of a four-stranded antiparallel beta sheet
The beta sheet, (β-sheet) (also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a g ...
with two alpha helices, is well conserved among all of these enzymes. In RdRp, the palm subdomain comprises three well-conserved motifs (A, B, and C). Motif A (D-x(4,5)-D) and motif C (GDD) are spatially juxtaposed; the aspartic acid
Aspartic acid (symbol Asp or D; the ionic form is known as aspartate), is an α-amino acid that is used in the biosynthesis of proteins. Like all other amino acids, it contains an amino group and a carboxylic acid. Its α-amino group is in the pro ...
residues of these motifs are implied in the binding of Mg2+ and/or Mn2+. The asparagine residue of motif B is involved in selection of ribonucleoside triphosphates over dNTPs and, thus, determines whether RNA rather than DNA is synthesized. The domain organization and the 3D structure of the catalytic centre of a wide range of RdRps, even those with a low overall sequence homology, are conserved. The catalytic center is formed by several motifs containing a number of conserved amino acid residues.
Eukaryotic RNA interference requires a cellular RNA-dependent RNA polymerase (c RdRp). Unlike the "hand" polymerases, they resemble simplified multi-subunit DNA-dependent RNA polymerases (DdRPs), specifically in the catalytic β/β' subunits, in that they use two sets of double-psi β-barrels in the active site. QDE1 () in '' Neurospora crassa'', which has both barrels in the same chain, is an example of such an c RdRp enzyme. Bacteriophage
A bacteriophage (), also known informally as a ''phage'' (), is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν ('), meaning "to devour". Bacteri ...
homologs of c RdRp, including the similarly single-chain DdRp yonO (), appear to be closer to c RdRps than DdRPs are.
In viruses
 There are 4 superfamilies of viruses that cover all RNA-containing viruses with no DNA stage:
* Viruses containing positive-strand RNA or double-strand RNA, except
There are 4 superfamilies of viruses that cover all RNA-containing viruses with no DNA stage:
* Viruses containing positive-strand RNA or double-strand RNA, except retrovirus
A retrovirus is a type of virus that inserts a DNA copy of its RNA genome into the DNA of a host cell that it invades, thus changing the genome of that cell. Once inside the host cell's cytoplasm, the virus uses its own reverse transcriptase ...
es and '' Birnaviridae''
** All positive-strand RNA eukaryotic viruses with no DNA stage
** All RNA-containing bacteriophage
A bacteriophage (), also known informally as a ''phage'' (), is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν ('), meaning "to devour". Bacteri ...
s; there are two families of RNA-containing bacteriophages: ''Leviviridae
''Fiersviridae'' is a family of positive-strand RNA viruses which infect prokaryotes. Bacteria serve as the natural host. They are small viruses with linear, positive-sense, single-stranded RNA genomes that encode four proteins. All phages of th ...
'' (positive ssRNA phages) and '' Cystoviridae'' (dsRNA phages)
** dsRNA virus family '' Reoviridae'', '' Totiviridae'', '' Hypoviridae'', '' Partitiviridae''
* '' Mononegavirales'' (negative-strand RNA viruses with non-segmented genomes; )
* Negative-strand RNA viruses with segmented genomes (), such as orthomyxoviruses and bunyaviruses
* dsRNA virus family '' Birnaviridae'' ()
Flaviviruses produce a polyprotein from the ssRNA genome. The polyprotein is cleaved to a number of products, one of which is NS5, an RNA-dependent RNA polymerase. This RNA-directed RNA polymerase possesses a number of short regions and motifs homologous to other RNA-directed RNA polymerases.
RNA replicase found in positive-strand ssRNA viruses are related to each other, forming three large superfamilies. Birnaviral RNA replicase is unique in that it lacks motif C (GDD) in the palm. Mononegaviral RdRp (PDB 5A22) has been automatically classified as similar to (+)−ssRNA RdRps, specifically one from ''Pestivirus'' and one from ''Leviviridae''. Bunyaviral RdRp monomer (PDB 5AMQ) resembles the heterotrimeric complex of Orthomyxoviral (Influenza; PDB 4WSB) RdRp.
Since it is a protein universal to RNA-containing viruses, RdRp is a useful marker for understanding their evolution. The overall structural evolution of viral RdRps has been reviewed.
Recombination
When replicating its (+)ssRNA genome, the poliovirus RdRp is able to carry out recombination. Recombination appears to occur by a copy choice mechanism in which the RdRp switches (+)ssRNA templates during negative strand synthesis. Recombination frequency is determined in part by the fidelity of RdRp replication. RdRp variants with high replication fidelity show reduced recombination, and low fidelity RdRps exhibit increased recombination. Recombination by RdRp strand switching also occurs frequently during replication in the (+)ssRNA plant carmoviruses and tombusviruses.Intragenic complementation
Sendai virus (family ''Paramyxoviridae'') has a linear, single stranded, negative-sense, nonsegmented RNA genome. The viral RdRp consists of two virus-encoded subunits, a smaller one P and a larger one L. When different inactive RdRp mutants with defects throughout the length of the L subunit where tested in pairwise combinations, restoration of viral RNA synthesis was observed in some combinations. This positive L–L interaction is referred to as intragenic complementation and indicates that the L protein is an oligomer in the viral RNA polymerase complex.Drug therapies
* RdRps can be used as drug targets for viral pathogens as their function is not necessary for eukaryotic survival. By inhibiting RNA-dependent RNA polymerase function, new RNAs cannot be replicated from an RNA template strand, however, DNA-dependent RNA polymerase will remain functional. * There are currently antiviral drugs against Hepatitis C and COVID-19 that specifically target RdRp. These includeSofosbuvir
Sofosbuvir, sold under the brand name Sovaldi among others, is a medication used to treat hepatitis C. It is taken Oral administration, by mouth.
Common side effects include fatigue, headache, nausea, and trouble sleeping. Side effects are gen ...
and Ribavirin against Hepatitis C and Remdesivir, the only FDA approved drug against COVID-19.
* GS-441524 triphosphate, is a substrate for RdRp, but not mammalian polymerases. It results in premature chain termination and inhibition of viral replication. GS-441524 triphosphate is the biologically active form of the phosphate pro-drug, Remdesivir. Remdesivir is classified as a nucleotide analog
Nucleoside analogues are nucleosides which contain a nucleic acid analogue and a sugar. Nucleotide analogs are nucleotides which contain a nucleic acid analogue, a sugar, and a phosphate group with one to three phosphates.
Nucleoside and nuc ...
in which it works to inhibit the function of RdRp by covalently binding to and interrupting termination of the nascent RNA through early or delayed termination or preventing further elongation of the RNA polynucleotide. This early termination leads to nonfunctional RNA that will be degraded through normal cellular processes.
RNA interference
The use of RNA-dependent RNA polymerase plays a major role in RNA interference in eukaryotes, a process used to silence gene expression via small interfering RNAs ( siRNAs) binding to mRNA rendering them inactive. Eukaryotic RdRp becomes active in the presence of dsRNA, and is a less widely distributed compared to other RNAi components as it lost in some animals, though still found in '' C. elegans'' and '' P. tetraurelia'' and plants. This presence of dsRNA triggers the activation of RdRp and RNAi processes by priming the initiation of RNA transcription through the introduction of siRNAs into the system. In ''C. elegans'', siRNAs are integrated into the RNA-induced silencing complex,RISC
In computer engineering, a reduced instruction set computer (RISC) is a computer designed to simplify the individual instructions given to the computer to accomplish tasks. Compared to the instructions given to a complex instruction set comput ...
, which works alongside mRNAs targeted for interference to recruit more RdRps to synthesize more secondary siRNAs and repress gene expression.
See also
*Spiegelman's Monster Spiegelman's Monster is the name given to an RNA chain of only 218 nucleotides that is able to be reproduced by the RNA replication enzyme RNA-dependent RNA polymerase, also called RNA replicase. It is named after its creator, Sol Spiegelman, of th ...
* NS5B inhibitor
Non-structural protein 5B (NS5B) inhibitors are a class of direct-acting antivirals widely used in the treatment of chronic hepatitis C. Depending on site of action and chemical composition, NS5B inh ...
Notes
References
External links
* * {{DEFAULTSORT:Rna-Dependent Rna Polymerase Gene expression RNA EC 2.7.7