RNA polymerase IV on:
[Wikipedia]
[Google]
[Amazon]
RNA polymerase IV (RNAP IV) is an
 Phylogenetic studies of land plants have led to the discovery of RNA
Phylogenetic studies of land plants have led to the discovery of RNA
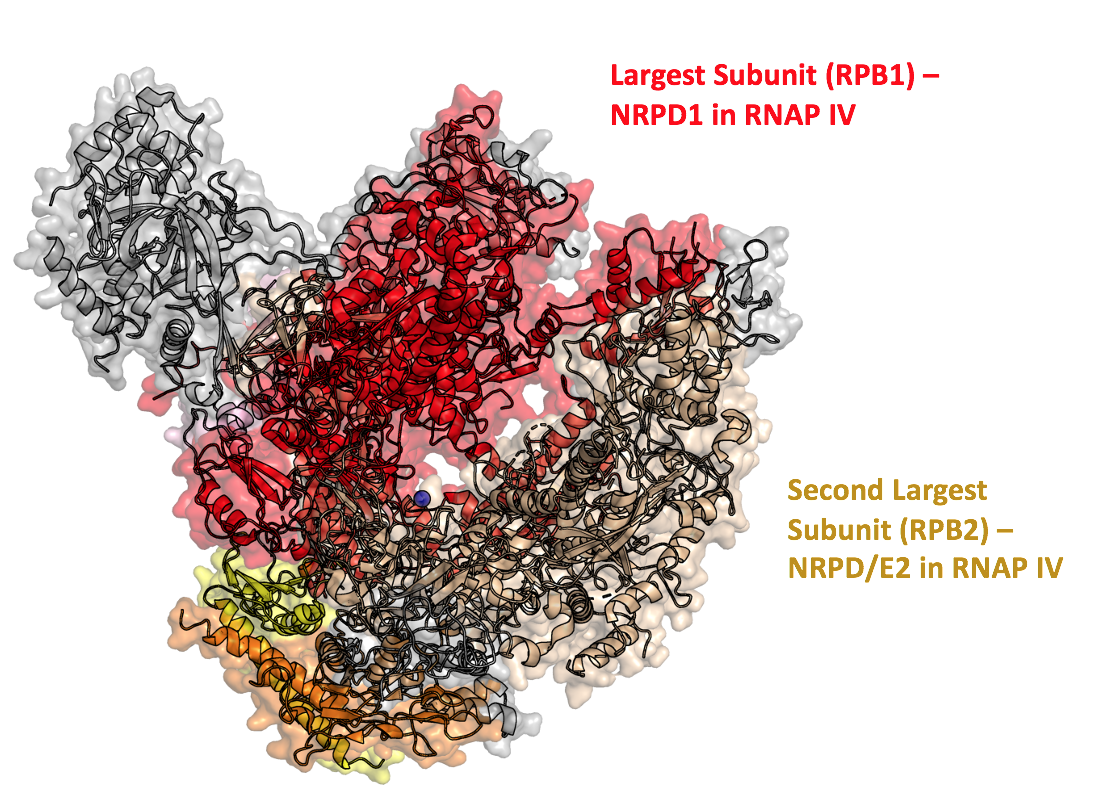
RPD1
and second-largest subunits
RPD2
of RNAP IV were analogous to the
 There is evidence that RNA Polymerase IV (RNAP IV) is responsible for producing
There is evidence that RNA Polymerase IV (RNAP IV) is responsible for producing
enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. A ...
that synthesizes small interfering RNA
Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of double-stranded RNA at first non-coding RNA molecules, typically 20-24 (normally 21) base pairs in length, similar to MicroRNA, miRNA, and op ...
(siRNA) in plants, which silence gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. The ...
. RNAP IV belongs to a family of enzymes that catalyze the process of transcription known as RNA Polymerases
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template.
Using the enzyme helicase, RNAP locally opens the ...
, which synthesize RNA from DNA templates. Discovered via phylogenetic
In biology, phylogenetics (; from Greek φυλή/ φῦλον [] "tribe, clan, race", and wikt:γενετικός, γενετικός [] "origin, source, birth") is the study of the evolutionary history and relationships among or within groups o ...
studies of land plants, genes of RNAP IV are thought to have resulted from multistep evolution processes that occurred in RNA Polymerase II
RNA polymerase II (RNAP II and Pol II) is a multiprotein complex that transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNAP enzymes found in the nucleus of eukaryoti ...
phylogenies. Such an evolutionary pathway is supported by the fact that RNAP IV is composed of 12 protein subunits that are either similar or identical to RNA polymerase II, and is specific to plant genomes. Via its synthesis of siRNA, RNAP IV is involved in regulation of heterochromatin
Heterochromatin is a tightly packed form of DNA or '' condensed DNA'', which comes in multiple varieties. These varieties lie on a continue between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a role ...
formation in a process known as RNA directed DNA Methylation (RdDM).
Discovery
Phylogenetic Studies
 Phylogenetic studies of land plants have led to the discovery of RNA
Phylogenetic studies of land plants have led to the discovery of RNA Polymerase
A polymerase is an enzyme ( EC 2.7.7.6/7/19/48/49) that synthesizes long chains of polymers or nucleic acids. DNA polymerase and RNA polymerase are used to assemble DNA and RNA molecules, respectively, by copying a DNA template strand using base- ...
IV. Analysis of the largestRPD1
and second-largest subunits
RPD2
of RNAP IV were analogous to the
Blast
Blast or The Blast may refer to:
*Explosion, a rapid increase in volume and release of energy in an extreme manner
*Detonation, an exothermic front accelerating through a medium that eventually drives a shock front
Film
* ''Blast'' (1997 film), ...
searches of RNAP II genes. Genes for RPD1 and RPD2 were found in all terrestrial plants, and the largest gene was found in the algal taxon, Charale. Further analysis of the origin of the protein indicates a gene duplication event of the largest subunit which suggested that the duplication event occurred after the divergence of Charales and land plants and algae. Specifically, the largest subunit in RNAP II formed RPD1 through a duplication event and the RPD2 gene arose due to a divergence. Evidence of these duplication events imply that the RNAP IV genes come from RNAP II phylogenies in a multistep process. In other words, the divergence
In vector calculus, divergence is a vector operator that operates on a vector field, producing a scalar field giving the quantity of the vector field's source at each point. More technically, the divergence represents the volume density of the ...
of the first subunit is the first step of multiple in the evolution of new RNAPs. RNAP IV also shares multiple subunits with RNAP II, in addition to the largest and second largest subunits, which was also suggested by continuous duplication events of particular lineages.
Distinction Between RNAP IV and RNAP V
Arabidopsis
''Arabidopsis'' (rockcress) is a genus in the family Brassicaceae. They are small flowering plants related to cabbage and mustard. This genus is of great interest since it contains thale cress (''Arabidopsis thaliana''), one of the model organi ...
expresses two forms of RNAP IV, formerly referred to as RNAP IVa and RNAP IVb, which differ at the largest subunit and have non redundant actions. Efficient silencing of transposons
A transposable element (TE, transposon, or jumping gene) is a nucleic acid sequence in DNA that can change its position within a genome, sometimes creating or reversing mutations and altering the cell's genetic identity and genome size. Tran ...
requires both RNAP IV forms while only RNAP IVa is required for basal silencing. This finding suggested the requirement of both forms for the mechanism of transposon methylation. Later experiments have shown that what was once thought to be two forms of RNAP IV are actually two structurally and functionally distinct polymerases. RNAP IVa was specified to be RNAP IV while RNAP IVb became known as RNAP V.
Structure
RNA Polymerase IV is composed of 12 protein subunits that are either similar or identical to the 12 subunits composingRNA Polymerase II
RNA polymerase II (RNAP II and Pol II) is a multiprotein complex that transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNAP enzymes found in the nucleus of eukaryoti ...
. Only four subunits distinguish RNAP IV structure from RNAP II and RNAP V. RNA Polymerase V differs from RNAP II by six subunits, indicating that both RNAP IV and RNAP V evolved from RNAP II in plants. In Arabidopsis
''Arabidopsis'' (rockcress) is a genus in the family Brassicaceae. They are small flowering plants related to cabbage and mustard. This genus is of great interest since it contains thale cress (''Arabidopsis thaliana''), one of the model organi ...
, two unique genes were found to encode subunits that distinguish RNAP IV from RNAP II. The largest subunit is encoded by NRPD1 (formerly NRPD1a), while the second largest subunit is encoded by NRPD2 and is shared with RNAP V. These subunits contain carboxyl-terminal domains (CTDs) which are necessary for the production of 20-30% of the siRNAs produced by RNA Polymerase IV, yet are not required for DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts t ...
.
Function
 There is evidence that RNA Polymerase IV (RNAP IV) is responsible for producing
There is evidence that RNA Polymerase IV (RNAP IV) is responsible for producing heterochromatin
Heterochromatin is a tightly packed form of DNA or '' condensed DNA'', which comes in multiple varieties. These varieties lie on a continue between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a role ...
, as dysfunction of either RNAP IV catalytic subunit (NRPD1 and NRPD2) disrupts the formation of heterochromatin. As heterochromatin is the silenced portion of DNA, it is formed when RNAP IV amplifies production of small interfering RNA
Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of double-stranded RNA at first non-coding RNA molecules, typically 20-24 (normally 21) base pairs in length, similar to MicroRNA, miRNA, and op ...
s (siRNA) that are responsible for methylating cytosine bases in DNA; this methylation silences segments of the genetic code, which can still be transcribed into mRNA but not translated into proteins. RNAP IV is involved in setting the methylation patterns in the 5S genes during plant maturation, resulting in the development of adult features in plants.
Mechanism
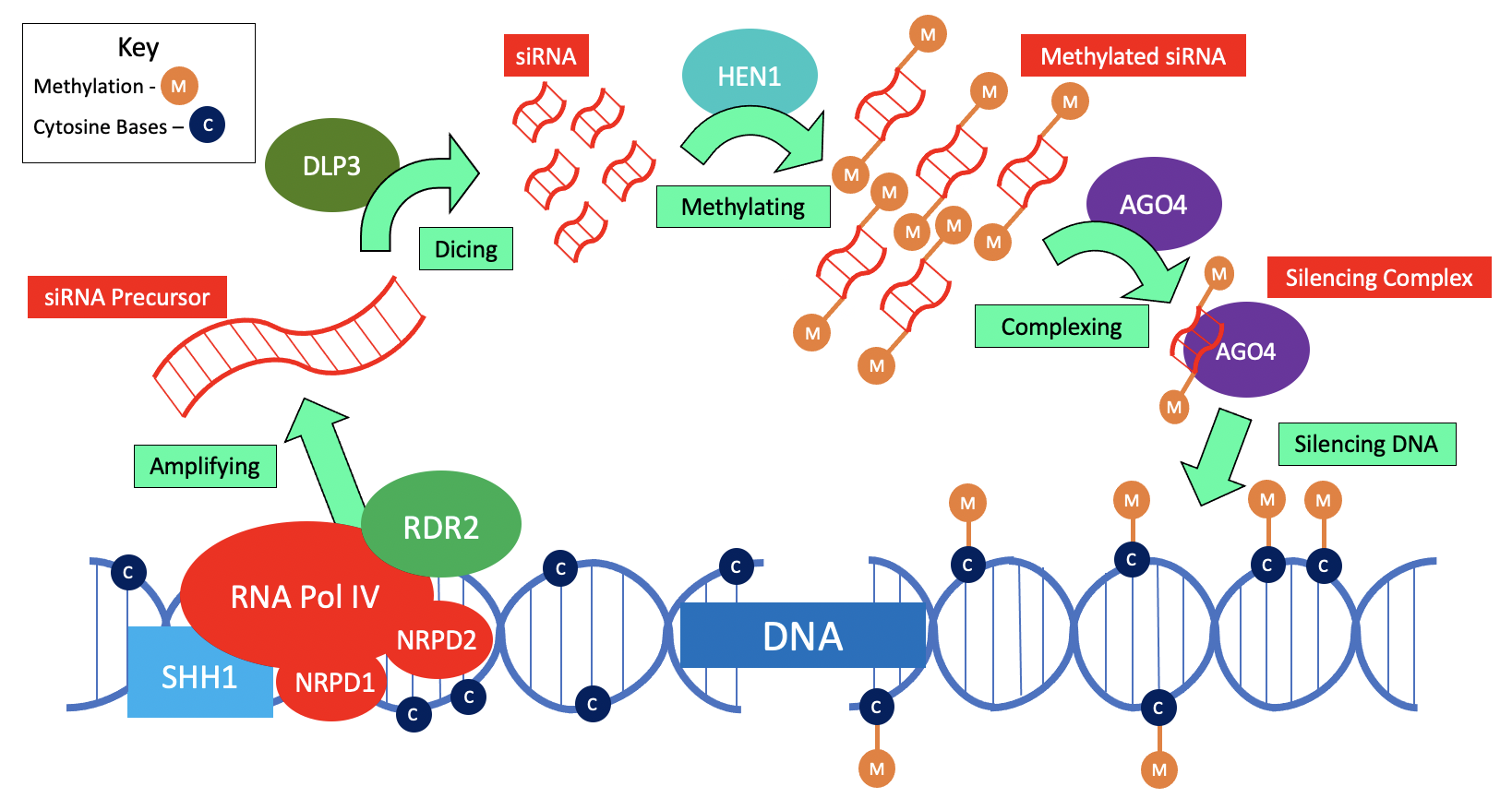
In the first step of heterochromatin formation, RNAP IV couples with anRNA-dependent RNA polymerase
RNA-dependent RNA polymerase (RdRp) or RNA replicase is an enzyme that catalyzes the replication of RNA from an RNA template. Specifically, it catalyzes synthesis of the RNA strand complementary to a given RNA template. This is in contrast to t ...
known as RDR2 to make a double stranded precursor to siRNA. Next, DICER-Like Protein 3 (DLP3), an enzyme which slices double stranded RNA substrates, cleaves the double stranded precursor into siRNAs that are each 24 nucleotides long. These siRNAs are then methylated at their 3’ ends by a protein known as HUA ENHANCER 1 (HEN1). Finally, these methylated siRNAs complex with a protein known as ARGONAUTE-4 (AGO4) in order to form the silencing complex that can perform the required methylation for heterochromatin production. This process is referred to as RNA-directed DNA Methylation
RNA-directed DNA methylation (RdDM) is a biological process in which non-coding RNA molecules direct the addition of DNA methylation to specific DNA sequences. The RdDM pathway is unique to plants, although other mechanisms of RNA-directed chromat ...
(RdDM) or Pol IV-mediated silencing as the introduction of these methyl groups by siRNAs silence both transposons
A transposable element (TE, transposon, or jumping gene) is a nucleic acid sequence in DNA that can change its position within a genome, sometimes creating or reversing mutations and altering the cell's genetic identity and genome size. Tran ...
and repetitive sequences of DNA.
Regulation
SAWADEE HOMEODOMAIN HOMOLOG 1 (SHH1) is a protein that interacts with RNAP IV and is critical in its regulation throughmethylation
In the chemical sciences, methylation denotes the addition of a methyl group on a substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These t ...
. SHH1 can only bind to chromatin
Chromatin is a complex of DNA and protein found in eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important roles in r ...
at specified “marked” segments, as its “SAWADEE” domain is a chromatin binding domain that probes for unmethylated K4 and methylated K9 modifications on the histone 3
Histone H3 is one of the five main histones involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'beads on a stri ...
(H3) tail of chromatin; its binding pocket
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate (binding site) a ...
s then attach to chromatin at these sites and allow RNAP IV occupancy at these same loci. In this manner, SHH1 functions to enable RNAP IV recruitment and stability at the most actively targeted genomic loci in RdDM in order to promote the previously mentioned siRNA
Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of double-stranded RNA at first non-coding RNA molecules, typically 20-24 (normally 21) base pairs in length, similar to miRNA, and operating ...
biogenesis of 24 nucleotide-long siRNA. Furthermore, it binds to repressive histone modifications, and any mutations that interfere with this process are associated with a reduction in DNA methylation and siRNA production. Regulation of siRNA production by RNAP IV through this mechanism results in major downstream effects, as the siRNAs produced in this manner defend the genome against the proliferation of invading viruses and endogenous transposable elements.
References
{{Polymerases Enzymes