Protein Patterns And Diagnosis on:
[Wikipedia]
[Google]
[Amazon]
 Proteins are large
Proteins are large
 With the development of X-ray crystallography, it became possible to sequence protein structures. The first protein structures to be solved were hemoglobin by Max Perutz and
With the development of X-ray crystallography, it became possible to sequence protein structures. The first protein structures to be solved were hemoglobin by Max Perutz and
 Most proteins consist of linear polymers built from series of up to 20 different L-α- amino acids. All proteinogenic amino acids possess common structural features, including an
Most proteins consist of linear polymers built from series of up to 20 different L-α- amino acids. All proteinogenic amino acids possess common structural features, including an

 Proteins are assembled from amino acids using information encoded in genes. Each protein has its own unique amino acid sequence that is specified by the nucleotide sequence of the gene encoding this protein. The genetic code is a set of three-nucleotide sets called
Proteins are assembled from amino acids using information encoded in genes. Each protein has its own unique amino acid sequence that is specified by the nucleotide sequence of the gene encoding this protein. The genetic code is a set of three-nucleotide sets called
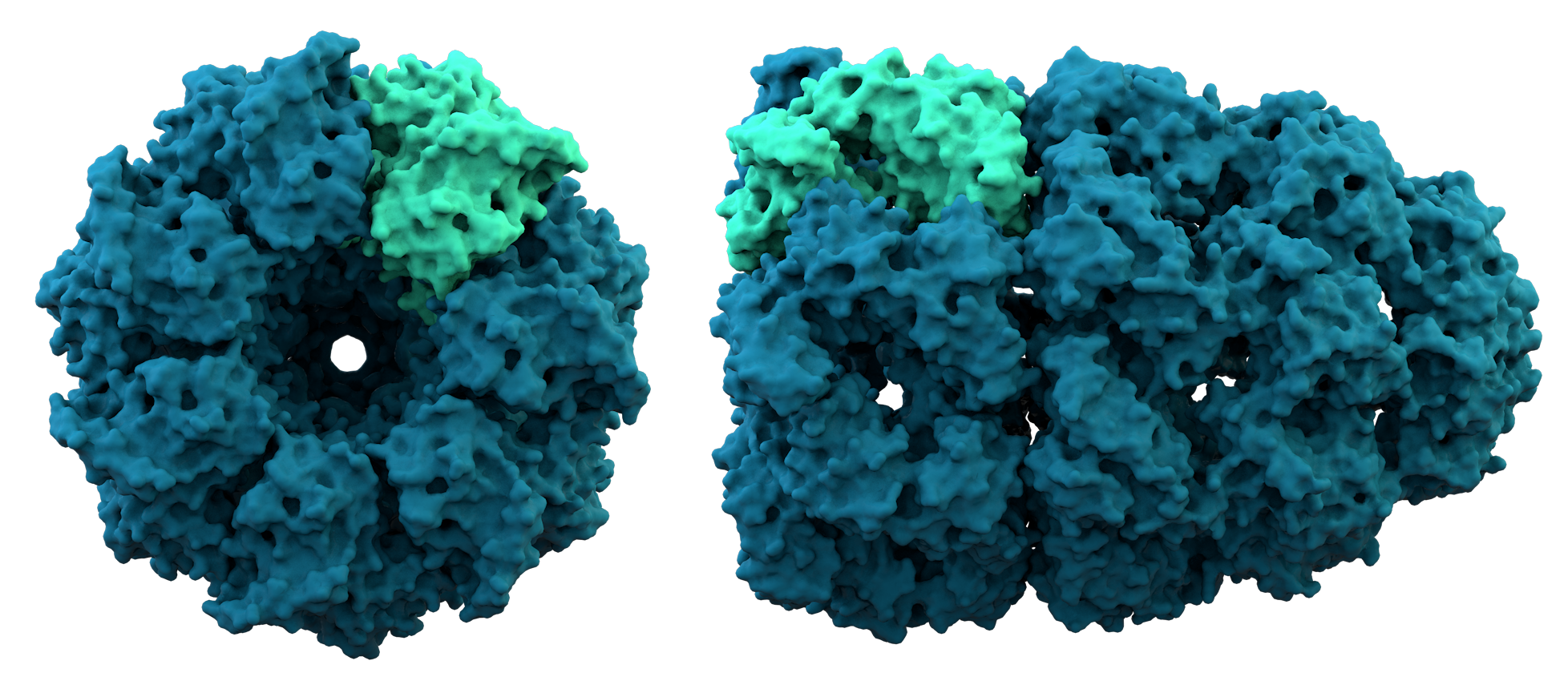 Most proteins
Most proteins  Proteins can be informally divided into three main classes, which correlate with typical tertiary structures: globular proteins, fibrous proteins, and membrane proteins. Almost all globular proteins are soluble and many are enzymes. Fibrous proteins are often structural, such as
Proteins can be informally divided into three main classes, which correlate with typical tertiary structures: globular proteins, fibrous proteins, and membrane proteins. Almost all globular proteins are soluble and many are enzymes. Fibrous proteins are often structural, such as
 The chief characteristic of proteins that also allows their diverse set of functions is their ability to bind other molecules specifically and tightly. The region of the protein responsible for binding another molecule is known as the binding site and is often a depression or "pocket" on the molecular surface. This binding ability is mediated by the tertiary structure of the protein, which defines the binding site pocket, and by the chemical properties of the surrounding amino acids' side chains. Protein binding can be extraordinarily tight and specific; for example, the ribonuclease inhibitor protein binds to human angiogenin with a sub-femtomolar
The chief characteristic of proteins that also allows their diverse set of functions is their ability to bind other molecules specifically and tightly. The region of the protein responsible for binding another molecule is known as the binding site and is often a depression or "pocket" on the molecular surface. This binding ability is mediated by the tertiary structure of the protein, which defines the binding site pocket, and by the chemical properties of the surrounding amino acids' side chains. Protein binding can be extraordinarily tight and specific; for example, the ribonuclease inhibitor protein binds to human angiogenin with a sub-femtomolar
 Many proteins are involved in the process of cell signaling and
Many proteins are involved in the process of cell signaling and
 The study of proteins ''in vivo'' is often concerned with the synthesis and localization of the protein within the cell. Although many intracellular proteins are synthesized in the cytoplasm and membrane-bound or secreted proteins in the
The study of proteins ''in vivo'' is often concerned with the synthesis and localization of the protein within the cell. Although many intracellular proteins are synthesized in the cytoplasm and membrane-bound or secreted proteins in the
 Complementary to the field of structural genomics, ''protein structure prediction'' develops efficient
Complementary to the field of structural genomics, ''protein structure prediction'' develops efficient
NCBI Entrez Protein database
NCBI Protein Structure database
Human Protein Reference Database
Human Proteinpedia
Folding@Home (Stanford University)
Protein Databank in Europe
(see als
PDBeQuips
short articles and tutorials on interesting PDB structures)
Research Collaboratory for Structural Bioinformatics
(see als
, presenting short accounts on selected proteins from the PDB)
Proteopedia – Life in 3D
rotatable, zoomable 3D model with wiki annotations for every known protein molecular structure.
UniProt the Universal Protein Resource
"An Introduction to Proteins"
from
Proteins: Biogenesis to Degradation – The Virtual Library of Biochemistry and Cell Biology
{{Authority control Molecular biology Proteomics
 Proteins are large
Proteins are large biomolecule
A biomolecule or biological molecule is a loosely used term for molecules present in organisms that are essential to one or more typically biological processes, such as cell division, morphogenesis, or development. Biomolecules include large ...
s and macromolecule
A macromolecule is a very large molecule important to biophysical processes, such as a protein or nucleic acid. It is composed of thousands of covalently bonded atoms. Many macromolecules are polymers of smaller molecules called monomers. The ...
s that comprise one or more long chains of amino acid residues
Residue may refer to:
Chemistry and biology
* An amino acid, within a peptide chain
* Crop residue, materials left after agricultural processes
* Pesticide residue, refers to the pesticides that may remain on or in food after they are appli ...
. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity.
A linear chain of amino acid residues is called a polypeptide
Peptides (, ) are short chains of amino acids linked by peptide bonds. Long chains of amino acids are called proteins. Chains of fewer than twenty amino acids are called oligopeptides, and include dipeptides, tripeptides, and tetrapeptides.
A p ...
. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bond
In organic chemistry, a peptide bond is an amide type of covalent chemical bond linking two consecutive alpha-amino acids from C1 (carbon number one) of one alpha-amino acid and N2 (nitrogen number two) of another, along a peptide or protein cha ...
s and adjacent amino acid residues. The sequence of amino acid residues in a protein is defined by the sequence of a gene, which is encoded in the genetic code. In general, the genetic code specifies 20 standard amino acids; but in certain organisms the genetic code can include selenocysteine
Selenocysteine (symbol Sec or U, in older publications also as Se-Cys) is the 21st proteinogenic amino acid. Selenoproteins contain selenocysteine residues. Selenocysteine is an analogue of the more common cysteine with selenium in place of the s ...
and—in certain archaea
Archaea ( ; singular archaeon ) is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria (in the Archaebac ...
—pyrrolysine
Pyrrolysine (symbol Pyl or O; encoded by the 'amber' stop codon UAG) is an α-amino acid that is used in the biosynthesis of proteins in some methanogenic archaea and bacteria; it is not present in humans. It contains an α-amino group (which is ...
. Shortly after or even during synthesis, the residues in a protein are often chemically modified by post-translational modification, which alters the physical and chemical properties, folding, stability, activity, and ultimately, the function of the proteins. Some proteins have non-peptide groups attached, which can be called prosthetic groups or cofactors
Cofactor may also refer to:
* Cofactor (biochemistry), a substance that needs to be present in addition to an enzyme for a certain reaction to be catalysed
* A domain parameter in elliptic curve cryptography, defined as the ratio between the order ...
. Proteins can also work together to achieve a particular function, and they often associate to form stable protein complexes.
Once formed, proteins only exist for a certain period and are then degraded and recycled by the cell's machinery through the process of protein turnover
In cell biology, protein turnover refers to the replacement of older proteins as they are broken down within the cell. Different types of proteins have very different turnover rates.
A balance between protein synthesis and protein degradation is ...
. A protein's lifespan is measured in terms of its half-life and covers a wide range. They can exist for minutes or years with an average lifespan of 1–2 days in mammalian cells. Abnormal or misfolded proteins are degraded more rapidly either due to being targeted for destruction or due to being unstable.
Like other biological macromolecules such as polysaccharide
Polysaccharides (), or polycarbohydrates, are the most abundant carbohydrates found in food. They are long chain polymeric carbohydrates composed of monosaccharide units bound together by glycosidic linkages. This carbohydrate can react with wa ...
s and nucleic acid
Nucleic acids are biopolymers, macromolecules, essential to all known forms of life. They are composed of nucleotides, which are the monomers made of three components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main cl ...
s, proteins are essential parts of organisms and participate in virtually every process within cells
Cell most often refers to:
* Cell (biology), the functional basic unit of life
Cell may also refer to:
Locations
* Monastic cell, a small room, hut, or cave in which a religious recluse lives, alternatively the small precursor of a monastery w ...
. Many proteins are enzymes that catalyse biochemical reactions and are vital to metabolism. Proteins also have structural or mechanical functions, such as actin and myosin
Myosins () are a superfamily of motor proteins best known for their roles in muscle contraction and in a wide range of other motility processes in eukaryotes. They are ATP-dependent and responsible for actin-based motility.
The first myosin ...
in muscle and the proteins in the cytoskeleton, which form a system of scaffolding that maintains cell shape. Other proteins are important in cell signaling, immune responses
An immune response is a reaction which occurs within an organism for the purpose of defending against foreign invaders. These invaders include a wide variety of different microorganisms including viruses, bacteria, parasites, and fungi which could ...
, cell adhesion
Cell adhesion is the process by which cells interact and attach to neighbouring cells through specialised molecules of the cell surface. This process can occur either through direct contact between cell surfaces such as cell junctions or indir ...
, and the cell cycle. In animals, proteins are needed in the diet to provide the essential amino acids that cannot be synthesized. Digestion breaks the proteins down for metabolic use.
Proteins may be purified from other cellular components using a variety of techniques such as ultracentrifugation, precipitation, electrophoresis
Electrophoresis, from Ancient Greek ἤλεκτρον (ḗlektron, "amber") and φόρησις (phórēsis, "the act of bearing"), is the motion of dispersed particles relative to a fluid under the influence of a spatially uniform electric fie ...
, and chromatography; the advent of genetic engineering
Genetic engineering, also called genetic modification or genetic manipulation, is the modification and manipulation of an organism's genes using technology. It is a set of technologies used to change the genetic makeup of cells, including t ...
has made possible a number of methods to facilitate purification. Methods commonly used to study protein structure and function include immunohistochemistry
Immunohistochemistry (IHC) is the most common application of immunostaining. It involves the process of selectively identifying antigens (proteins) in cells of a tissue section by exploiting the principle of antibodies binding specifically to an ...
, site-directed mutagenesis, X-ray crystallography, nuclear magnetic resonance and mass spectrometry
Mass spectrometry (MS) is an analytical technique that is used to measure the mass-to-charge ratio of ions. The results are presented as a ''mass spectrum'', a plot of intensity as a function of the mass-to-charge ratio. Mass spectrometry is use ...
.
History and etymology
Proteins were recognized as a distinct class of biological molecules in the eighteenth century by Antoine Fourcroy and others, distinguished by the molecules' ability to coagulate or flocculate under treatments with heat or acid. Noted examples at the time included albumin fromegg white
Egg white is the clear liquid (also called the albumen or the glair/glaire) contained within an egg. In chickens it is formed from the layers of secretions of the anterior section of the hen's oviduct during the passage of the egg. It forms arou ...
s, blood serum albumin, fibrin
Fibrin (also called Factor Ia) is a fibrous, non-globular protein involved in the clotting of blood. It is formed by the action of the protease thrombin on fibrinogen, which causes it to polymerize. The polymerized fibrin, together with platele ...
, and wheat gluten.
Proteins were first described by the Dutch chemist Gerardus Johannes Mulder
Gerardus Johannes Mulder or Gerrit Jan Mulder (27 December 1802 – 18 April 1880) was a Dutch organic and analytical chemist.
Life
Mulder was born in Utrecht and earned a medical degree from Utrecht University.
He became a reader of chemis ...
and named by the Swedish chemist Jöns Jacob Berzelius
Baron Jöns Jacob Berzelius (; by himself and his contemporaries named only Jacob Berzelius, 20 August 1779 – 7 August 1848) was a Swedish chemist. Berzelius is considered, along with Robert Boyle, John Dalton, and Antoine Lavoisier, to be on ...
in 1838. Mulder carried out elemental analysis
Elemental analysis is a process where a sample of some material (e.g., soil, waste or drinking water, bodily fluids, minerals, chemical compounds) is analyzed for its elemental and sometimes isotopic composition. Elemental analysis can be qualita ...
of common proteins and found that nearly all proteins had the same empirical formula, C400H620N100O120P1S1. He came to the erroneous conclusion that they might be composed of a single type of (very large) molecule. The term "protein" to describe these molecules was proposed by Mulder's associate Berzelius; protein is derived from the Greek word (), meaning "primary", "in the lead", or "standing in front", + '' -in''. Mulder went on to identify the products of protein degradation such as the amino acid leucine for which he found a (nearly correct) molecular weight of 131 Da. Prior to "protein", other names were used, like "albumins" or "albuminous materials" (''Eiweisskörper'', in German).
Early nutritional scientists such as the German Carl von Voit
Carl von Voit (31 October 1831 – 31 January 1908) was a German physiologist and dietitian.
Biography
Voit was born in Amberg, the son of August von Voit and Mathilde Burgett. From 1848 to 1854 he studied at the universities of Munich and Wür ...
believed that protein was the most important nutrient for maintaining the structure of the body, because it was generally believed that "flesh makes flesh." Karl Heinrich Ritthausen
Karl Heinrich Ritthausen (13 January 1826 – 16 October 1912) was a German biochemist who identified two amino acids and made other contributions to the science of plant proteins.
Education
Ritthausen was born in Armenruh, near Goldburg, Sil ...
extended known protein forms with the identification of glutamic acid
Glutamic acid (symbol Glu or E; the ionic form is known as glutamate) is an α-amino acid that is used by almost all living beings in the biosynthesis of proteins. It is a non-essential nutrient for humans, meaning that the human body can synt ...
. At the Connecticut Agricultural Experiment Station a detailed review of the vegetable proteins was compiled by Thomas Burr Osborne. Working with Lafayette Mendel and applying Liebig's law of the minimum in feeding laboratory rats, the nutritionally essential amino acids were established. The work was continued and communicated by William Cumming Rose. The understanding of proteins as polypeptide
Peptides (, ) are short chains of amino acids linked by peptide bonds. Long chains of amino acids are called proteins. Chains of fewer than twenty amino acids are called oligopeptides, and include dipeptides, tripeptides, and tetrapeptides.
A p ...
s came through the work of Franz Hofmeister
Franz Hofmeister (30 August 1850, in Prague – 26 July 1922, in Würzburg) was an early protein scientist, and is famous for his studies of salts that influence the solubility and conformational stability of proteins. In 1902, Hofmeister became t ...
and Hermann Emil Fischer
Hermann Emil Louis Fischer (; 9 October 1852 – 15 July 1919) was a German chemist and 1902 recipient of the Nobel Prize in Chemistry. He discovered the Fischer esterification. He also developed the Fischer projection, a symbolic way of dra ...
in 1902. The central role of proteins as enzymes in living organisms was not fully appreciated until 1926, when James B. Sumner
James Batcheller Sumner (November 19, 1887 – August 12, 1955) was an American chemist. He discovered that enzymes can be crystallized, for which he shared the Nobel Prize in Chemistry in 1946 with John Howard Northrop and Wendell Meredith Stanl ...
showed that the enzyme urease was in fact a protein.
The difficulty in purifying proteins in large quantities made them very difficult for early protein biochemists to study. Hence, early studies focused on proteins that could be purified in large quantities, e.g., those of blood, egg white, various toxins, and digestive/metabolic enzymes obtained from slaughterhouses. In the 1950s, the Armour Hot Dog Co. purified 1 kg of pure bovine pancreatic ribonuclease A and made it freely available to scientists; this gesture helped ribonuclease A become a major target for biochemical study for the following decades.
Linus Pauling
Linus Carl Pauling (; February 28, 1901August 19, 1994) was an American chemist, biochemist, chemical engineer, peace activist, author, and educator. He published more than 1,200 papers and books, of which about 850 dealt with scientific top ...
is credited with the successful prediction of regular protein secondary structure
Protein secondary structure is the three dimensional conformational isomerism, form of ''local segments'' of proteins. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta ...
s based on hydrogen bonding, an idea first put forth by William Astbury in 1933. Later work by Walter Kauzmann
Walter J. Kauzmann (18 August 1916 – 27 January 2009) was an American chemist and professor emeritus of Princeton University. He was noted for his work in both physical chemistry and biochemistry. His most important contribution was recognizin ...
on denaturation, based partly on previous studies by Kaj Linderstrøm-Lang, contributed an understanding of protein folding and structure mediated by hydrophobic interactions
The hydrophobic effect is the observed tendency of nonpolar substances to aggregate in an aqueous solution and exclude water molecules. The word hydrophobic literally means "water-fearing", and it describes the segregation of water and nonpolar ...
.
The first protein to be sequenced was insulin
Insulin (, from Latin ''insula'', 'island') is a peptide hormone produced by beta cells of the pancreatic islets encoded in humans by the ''INS'' gene. It is considered to be the main anabolic hormone of the body. It regulates the metabolism o ...
, by Frederick Sanger
Frederick Sanger (; 13 August 1918 – 19 November 2013) was an English biochemist who received the Nobel Prize in Chemistry twice.
He won the 1958 Chemistry Prize for determining the amino acid sequence of insulin and numerous other p ...
, in 1949. Sanger correctly determined the amino acid sequence of insulin, thus conclusively demonstrating that proteins consisted of linear polymers of amino acids rather than branched chains, colloid
A colloid is a mixture in which one substance consisting of microscopically dispersed insoluble particles is suspended throughout another substance. Some definitions specify that the particles must be dispersed in a liquid, while others extend ...
s, or cyclol
The cyclol hypothesis is the now discredited first structural model of a folded, globular protein, formulated in the 1930s. It was based on the cyclol reaction of peptide bonds proposed by physicist Frederick Frank in 1936, in which two pe ...
s. He won the Nobel Prize for this achievement in 1958.
 With the development of X-ray crystallography, it became possible to sequence protein structures. The first protein structures to be solved were hemoglobin by Max Perutz and
With the development of X-ray crystallography, it became possible to sequence protein structures. The first protein structures to be solved were hemoglobin by Max Perutz and myoglobin
Myoglobin (symbol Mb or MB) is an iron- and oxygen-binding protein found in the cardiac and skeletal muscle tissue of vertebrates in general and in almost all mammals. Myoglobin is distantly related to hemoglobin. Compared to hemoglobin, myoglobi ...
by John Kendrew, in 1958. The use of computers and increasing computing power also supported the sequencing of complex proteins. In 1999, Roger Kornberg succeeded in sequencing the highly complex structure of RNA polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template.
Using the enzyme helicase, RNAP locally opens the ...
using high intensity X-rays from synchrotrons
A synchrotron is a particular type of cyclic particle accelerator, descended from the cyclotron, in which the accelerating particle beam travels around a fixed closed-loop path. The magnetic field which bends the particle beam into its closed p ...
.
Since then, cryo-electron microscopy (cryo-EM) of large macromolecular assemblies
The term macromolecular assembly (MA) refers to massive chemical structures such as viruses and non-biologic nanoparticles, cellular organelles and membranes and ribosomes, etc. that are complex mixtures of polypeptide, polynucleotide, polysaccha ...
has been developed. Cryo-EM uses protein samples that are frozen rather than crystals, and beams of electrons rather than x-rays. It causes less damage to the sample, allowing scientists to obtain more information and analyze larger structures. Computational protein structure prediction of small protein domains has also helped researchers to approach atomic-level resolution of protein structures.
, the Protein Data Bank has over 126,060 atomic-resolution structures of proteins.
Number of proteins encoded in genomes
The number of proteins encoded in a genome roughly corresponds to the number of genes (although there may be a significant number of genes that encodeRNA
Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and deoxyribonucleic acid ( DNA) are nucleic acids. Along with lipids, proteins, and carbohydra ...
of protein, e.g. ribosomal RNAs). Viruses typically encode a few to a few hundred proteins, archaea
Archaea ( ; singular archaeon ) is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria (in the Archaebac ...
and bacteria a few hundred to a few thousand, while eukaryote
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacte ...
s typically encode a few thousand up to tens of thousands of proteins (see genome size for a list of examples).
Biochemistry
 Most proteins consist of linear polymers built from series of up to 20 different L-α- amino acids. All proteinogenic amino acids possess common structural features, including an
Most proteins consist of linear polymers built from series of up to 20 different L-α- amino acids. All proteinogenic amino acids possess common structural features, including an α-carbon
In the nomenclature of organic chemistry, a locant is a term to indicate the position of a functional group or substituent within a molecule.
Numeric locants
The International Union of Pure and Applied Chemistry (IUPAC) recommends the use of ...
to which an amino group, a carboxyl group, and a variable side chain are bonded. Only proline
Proline (symbol Pro or P) is an organic acid classed as a proteinogenic amino acid (used in the biosynthesis of proteins), although it does not contain the amino group but is rather a secondary amine. The secondary amine nitrogen is in the prot ...
differs from this basic structure as it contains an unusual ring to the N-end amine group, which forces the CO–NH amide moiety into a fixed conformation. The side chains of the standard amino acids, detailed in the list of standard amino acids
Proteinogenic amino acids are amino acids that are incorporated biosynthetically into proteins during translation. The word "proteinogenic" means "protein creating". Throughout known life, there are 22 genetically encoded (proteinogenic) amino aci ...
, have a great variety of chemical structures and properties; it is the combined effect of all of the amino acid side chains in a protein that ultimately determines its three-dimensional structure and its chemical reactivity.
The amino acids in a polypeptide chain are linked by peptide bond
In organic chemistry, a peptide bond is an amide type of covalent chemical bond linking two consecutive alpha-amino acids from C1 (carbon number one) of one alpha-amino acid and N2 (nitrogen number two) of another, along a peptide or protein cha ...
s. Once linked in the protein chain, an individual amino acid is called a ''residue,'' and the linked series of carbon, nitrogen, and oxygen atoms are known as the ''main chain'' or ''protein backbone.''
The peptide bond has two resonance forms that contribute some double-bond
In chemistry, a double bond is a covalent bond between two atoms involving four bonding electrons as opposed to two in a single bond. Double bonds occur most commonly between two carbon atoms, for example in alkenes. Many double bonds exist betw ...
character and inhibit rotation around its axis, so that the alpha carbons are roughly coplanar. The other two dihedral angles in the peptide bond determine the local shape assumed by the protein backbone. The end with a free amino group is known as the N-terminus
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the ami ...
or amino terminus, whereas the end of the protein with a free carboxyl group is known as the C-terminus
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is ...
or carboxy terminus (the sequence of the protein is written from N-terminus to C-terminus, from left to right).
The words ''protein'', ''polypeptide,'' and '' peptide'' are a little ambiguous and can overlap in meaning. ''Protein'' is generally used to refer to the complete biological molecule in a stable conformation, whereas ''peptide'' is generally reserved for a short amino acid oligomers often lacking a stable 3D structure. But the boundary between the two is not well defined and usually lies near 20–30 residues. ''Polypeptide'' can refer to any single linear chain of amino acids, usually regardless of length, but often implies an absence of a defined conformation.
Interactions
Proteins can interact with many types of molecules, including with other proteins, with lipids, with carbohydrates, and with DNA.Abundance in cells
It has been estimated that average-sized bacteria contain about 2 million proteins per cell (e.g. ''E. coli
''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus ''Escher ...
'' and ''Staphylococcus aureus
''Staphylococcus aureus'' is a Gram-positive spherically shaped bacterium, a member of the Bacillota, and is a usual member of the microbiota of the body, frequently found in the upper respiratory tract and on the skin. It is often positive ...
''). Smaller bacteria, such as ''Mycoplasma
''Mycoplasma'' is a genus of bacteria that, like the other members of the class ''Mollicutes'', lack a cell wall around their cell membranes. Peptidoglycan (murein) is absent. This characteristic makes them naturally resistant to antibiotics ...
'' or ''spirochetes
A spirochaete () or spirochete is a member of the phylum Spirochaetota (), (synonym Spirochaetes) which contains distinctive diderm (double-membrane) gram-negative bacteria, most of which have long, helically coiled (corkscrew-shaped or s ...
'' contain fewer molecules, on the order of 50,000 to 1 million. By contrast, eukaryotic cells are larger and thus contain much more protein. For instance, yeast cells have been estimated to contain about 50 million proteins and human cells on the order of 1 to 3 billion. The concentration of individual protein copies ranges from a few molecules per cell up to 20 million. Not all genes coding proteins are expressed in most cells and their number depends on, for example, cell type and external stimuli. For instance, of the 20,000 or so proteins encoded by the human genome, only 6,000 are detected in lymphoblastoid
__NOTOC__
A lymphoblast is a modified naive lymphocyte with altered cell morphology. It occurs when the lymphocyte is activated by an antigen (from antigen-presenting cells) and increased in volume by nucleus and cytoplasm growth as well as new mRN ...
cells.
Synthesis
Biosynthesis
codon
The genetic code is the set of rules used by living cells to translate information encoded within genetic material ( DNA or RNA sequences of nucleotide triplets, or codons) into proteins. Translation is accomplished by the ribosome, which links ...
s and each three-nucleotide combination designates an amino acid, for example AUG ( adenine– uracil– guanine) is the code for methionine
Methionine (symbol Met or M) () is an essential amino acid in humans. As the precursor of other amino acids such as cysteine and taurine, versatile compounds such as SAM-e, and the important antioxidant glutathione, methionine plays a critical ro ...
. Because DNA contains four nucleotides, the total number of possible codons is 64; hence, there is some redundancy in the genetic code, with some amino acids specified by more than one codon. Genes encoded in DNA are first transcribed into pre-messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the p ...
(mRNA) by proteins such as RNA polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template.
Using the enzyme helicase, RNAP locally opens the ...
. Most organisms then process the pre-mRNA (also known as a ''primary transcript'') using various forms of Post-transcriptional modification to form the mature mRNA, which is then used as a template for protein synthesis by the ribosome
Ribosomes ( ) are macromolecular machines, found within all cells, that perform biological protein synthesis (mRNA translation). Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to ...
. In prokaryotes the mRNA may either be used as soon as it is produced, or be bound by a ribosome after having moved away from the nucleoid
The nucleoid (meaning ''nucleus-like'') is an irregularly shaped region within the prokaryotic cell that contains all or most of the genetic material. The chromosome of a prokaryote is circular, and its length is very large compared to the cell dim ...
. In contrast, eukaryote
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacte ...
s make mRNA in the cell nucleus
The cell nucleus (pl. nuclei; from Latin or , meaning ''kernel'' or ''seed'') is a membrane-bound organelle found in eukaryotic cells. Eukaryotic cells usually have a single nucleus, but a few cell types, such as mammalian red blood cells, h ...
and then translocate it across the nuclear membrane into the cytoplasm, where protein synthesis
Protein biosynthesis (or protein synthesis) is a core biological process, occurring inside Cell (biology), cells, homeostasis, balancing the loss of cellular proteins (via Proteolysis, degradation or Protein targeting, export) through the product ...
then takes place. The rate of protein synthesis is higher in prokaryotes than eukaryotes and can reach up to 20 amino acids per second.
The process of synthesizing a protein from an mRNA template is known as translation. The mRNA is loaded onto the ribosome and is read three nucleotides at a time by matching each codon to its base pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
ing anticodon located on a transfer RNA
Transfer RNA (abbreviated tRNA and formerly referred to as sRNA, for soluble RNA) is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes), that serves as the physical link between the mRNA and the amino ac ...
molecule, which carries the amino acid corresponding to the codon it recognizes. The enzyme aminoacyl tRNA synthetase "charges" the tRNA molecules with the correct amino acids. The growing polypeptide is often termed the ''nascent chain''. Proteins are always biosynthesized from N-terminus
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the ami ...
to C-terminus
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is ...
.
The size of a synthesized protein can be measured by the number of amino acids it contains and by its total molecular mass, which is normally reported in units of ''daltons'' (synonymous with atomic mass units), or the derivative unit kilodalton (kDa). The average size of a protein increases from Archaea to Bacteria to Eukaryote (283, 311, 438 residues and 31, 34, 49 kDa respectively) due to a bigger number of protein domains constituting proteins in higher organisms. For instance, yeast proteins are on average 466 amino acids long and 53 kDa in mass. The largest known proteins are the titins, a component of the muscle
Skeletal muscles (commonly referred to as muscles) are organs of the vertebrate muscular system and typically are attached by tendons to bones of a skeleton. The muscle cells of skeletal muscles are much longer than in the other types of muscl ...
sarcomere, with a molecular mass of almost 3,000 kDa and a total length of almost 27,000 amino acids.
Chemical synthesis
Short proteins can also be synthesized chemically by a family of methods known as peptide synthesis, which rely onorganic synthesis
Organic synthesis is a special branch of chemical synthesis and is concerned with the intentional construction of organic compounds. Organic molecules are often more complex than inorganic compounds, and their synthesis has developed into one o ...
techniques such as chemical ligation
Chemical ligation is a set of techniques used for creating long peptide or protein chains. It is the second step of a convergent approach. First, smaller peptides containing 30-50 amino acids are prepared by conventional chemical peptide synthe ...
to produce peptides in high yield. Chemical synthesis allows for the introduction of non-natural amino acids into polypeptide chains, such as attachment of fluorescent probes to amino acid side chains. These methods are useful in laboratory biochemistry and cell biology
Cell biology (also cellular biology or cytology) is a branch of biology that studies the structure, function, and behavior of cells. All living organisms are made of cells. A cell is the basic unit of life that is responsible for the living and ...
, though generally not for commercial applications. Chemical synthesis is inefficient for polypeptides longer than about 300 amino acids, and the synthesized proteins may not readily assume their native tertiary structure. Most chemical synthesis methods proceed from C-terminus to N-terminus, opposite the biological reaction.
Structure
 Most proteins
Most proteins fold
Fold, folding or foldable may refer to:
Arts, entertainment, and media
* ''Fold'' (album), the debut release by Australian rock band Epicure
*Fold (poker), in the game of poker, to discard one's hand and forfeit interest in the current pot
*Above ...
into unique 3D structures. The shape into which a protein naturally folds is known as its native conformation
In biochemistry, the native state of a protein or nucleic acid is its properly folded and/or assembled form, which is operative and functional. The native state of a biomolecule may possess all four levels of biomolecular structure, with the ...
. Although many proteins can fold unassisted, simply through the chemical properties of their amino acids, others require the aid of molecular chaperones to fold into their native states. Biochemists often refer to four distinct aspects of a protein's structure:
* ''Primary structure
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthes ...
'': the amino acid sequence. A protein is a polyamide.
* ''Secondary structure
Protein secondary structure is the three dimensional conformational isomerism, form of ''local segments'' of proteins. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta ...
'': regularly repeating local structures stabilized by hydrogen bond
In chemistry, a hydrogen bond (or H-bond) is a primarily electrostatic force of attraction between a hydrogen (H) atom which is covalently bound to a more electronegative "donor" atom or group (Dn), and another electronegative atom bearing a ...
s. The most common examples are the α-helix, β-sheet and turns. Because secondary structures are local, many regions of different secondary structure can be present in the same protein molecule.
* '' Tertiary structure'': the overall shape of a single protein molecule; the spatial relationship of the secondary structures to one another. Tertiary structure is generally stabilized by nonlocal interactions, most commonly the formation of a hydrophobic core, but also through salt bridges, hydrogen bonds, disulfide bonds, and even posttranslational modifications. The term "tertiary structure" is often used as synonymous with the term ''fold''. The tertiary structure is what controls the basic function of the protein.
* '' Quaternary structure'': the structure formed by several protein molecules (polypeptide chains), usually called ''protein subunit
In structural biology, a protein subunit is a polypeptide chain or single protein molecule that assembles (or "''coassembles''") with others to form a protein complex.
Large assemblies of proteins such as viruses often use a small number of ty ...
s'' in this context, which function as a single protein complex.
* '' Quinary structure'': the signatures of protein surface that organize the crowded cellular interior. Quinary structure is dependent on transient, yet essential, macromolecular interactions that occur inside living cells.
Proteins are not entirely rigid molecules. In addition to these levels of structure, proteins may shift between several related structures while they perform their functions. In the context of these functional rearrangements, these tertiary or quaternary structures are usually referred to as " conformations", and transitions between them are called ''conformational changes.'' Such changes are often induced by the binding of a substrate
Substrate may refer to:
Physical layers
*Substrate (biology), the natural environment in which an organism lives, or the surface or medium on which an organism grows or is attached
** Substrate (locomotion), the surface over which an organism lo ...
molecule to an enzyme's active site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate (binding site) a ...
, or the physical region of the protein that participates in chemical catalysis. In solution proteins also undergo variation in structure through thermal vibration and the collision with other molecules.
 Proteins can be informally divided into three main classes, which correlate with typical tertiary structures: globular proteins, fibrous proteins, and membrane proteins. Almost all globular proteins are soluble and many are enzymes. Fibrous proteins are often structural, such as
Proteins can be informally divided into three main classes, which correlate with typical tertiary structures: globular proteins, fibrous proteins, and membrane proteins. Almost all globular proteins are soluble and many are enzymes. Fibrous proteins are often structural, such as collagen
Collagen () is the main structural protein in the extracellular matrix found in the body's various connective tissues. As the main component of connective tissue, it is the most abundant protein in mammals, making up from 25% to 35% of the whole ...
, the major component of connective tissue, or keratin, the protein component of hair and nails. Membrane proteins often serve as receptors or provide channels for polar or charged molecules to pass through the cell membrane.
A special case of intramolecular hydrogen bonds within proteins, poorly shielded from water attack and hence promoting their own dehydration, are called dehydrons.
Protein domains
Many proteins are composed of several protein domains, i.e. segments of a protein that fold into distinct structural units. Domains usually also have specific functions, such as enzymatic activities (e.g.kinase
In biochemistry, a kinase () is an enzyme that catalyzes the transfer of phosphate groups from high-energy, phosphate-donating molecules to specific substrates. This process is known as phosphorylation, where the high-energy ATP molecule don ...
) or they serve as binding modules (e.g. the SH3 domain binds to proline-rich sequences in other proteins).
Sequence motif
Short amino acid sequences within proteins often act as recognition sites for other proteins. For instance, SH3 domains typically bind to short PxxP motifs (i.e. 2proline
Proline (symbol Pro or P) is an organic acid classed as a proteinogenic amino acid (used in the biosynthesis of proteins), although it does not contain the amino group but is rather a secondary amine. The secondary amine nitrogen is in the prot ...
s separated by two unspecified amino acids although the surrounding amino acids may determine the exact binding specificity). Many such motifs has been collected in the Eukaryotic Linear Motif (ELM) database.
Cellular functions
Proteins are the chief actors within the cell, said to be carrying out the duties specified by the information encoded in genes. With the exception of certain types ofRNA
Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and deoxyribonucleic acid ( DNA) are nucleic acids. Along with lipids, proteins, and carbohydra ...
, most other biological molecules are relatively inert elements upon which proteins act. Proteins make up half the dry weight of an '' Escherichia coli'' cell, whereas other macromolecules such as DNA and RNA make up only 3% and 20%, respectively.Voet D, Voet JG. (2004). ''Biochemistry'' Vol 1 3rd ed. Wiley: Hoboken, NJ. The set of proteins expressed in a particular cell or cell type is known as its proteome.
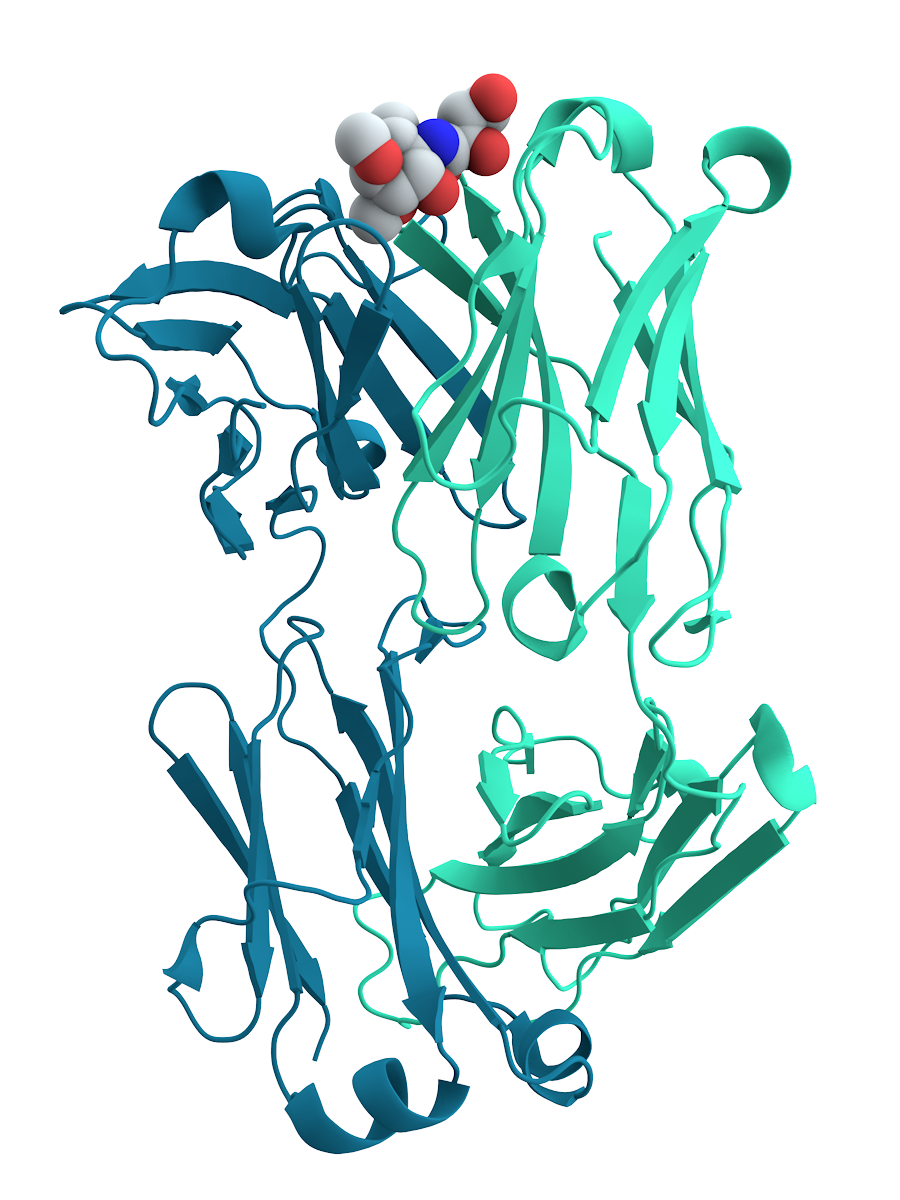
 The chief characteristic of proteins that also allows their diverse set of functions is their ability to bind other molecules specifically and tightly. The region of the protein responsible for binding another molecule is known as the binding site and is often a depression or "pocket" on the molecular surface. This binding ability is mediated by the tertiary structure of the protein, which defines the binding site pocket, and by the chemical properties of the surrounding amino acids' side chains. Protein binding can be extraordinarily tight and specific; for example, the ribonuclease inhibitor protein binds to human angiogenin with a sub-femtomolar
The chief characteristic of proteins that also allows their diverse set of functions is their ability to bind other molecules specifically and tightly. The region of the protein responsible for binding another molecule is known as the binding site and is often a depression or "pocket" on the molecular surface. This binding ability is mediated by the tertiary structure of the protein, which defines the binding site pocket, and by the chemical properties of the surrounding amino acids' side chains. Protein binding can be extraordinarily tight and specific; for example, the ribonuclease inhibitor protein binds to human angiogenin with a sub-femtomolar dissociation constant
In chemistry, biochemistry, and pharmacology, a dissociation constant (K_D) is a specific type of equilibrium constant that measures the propensity of a larger object to separate (dissociate) reversibly into smaller components, as when a complex fa ...
(<10−15 M) but does not bind at all to its amphibian homolog onconase
Ranpirnase is a ribonuclease enzyme found in the oocytes of the Northern Leopard Frog (''Rana pipiens''). Ranpirnase is a member of the pancreatic ribonuclease (RNase A) protein superfamily and degrades RNA substrates with a sequence preference fo ...
(>1 M). Extremely minor chemical changes such as the addition of a single methyl group to a binding partner can sometimes suffice to nearly eliminate binding; for example, the aminoacyl tRNA synthetase specific to the amino acid valine discriminates against the very similar side chain of the amino acid isoleucine
Isoleucine (symbol Ile or I) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form under biological conditions), an α-carboxylic acid group (which is in the deprot ...
.
Proteins can bind to other proteins as well as to small-molecule
Within the fields of molecular biology and pharmacology, a small molecule or micromolecule is a low molecular weight (≤ 1000 daltons) organic compound that may regulate a biological process, with a size on the order of 1 nm. Many drugs ar ...
substrates. When proteins bind specifically to other copies of the same molecule, they can oligomer
In chemistry and biochemistry, an oligomer () is a molecule that consists of a few repeating units which could be derived, actually or conceptually, from smaller molecules, monomers.Quote: ''Oligomer molecule: A molecule of intermediate relativ ...
ize to form fibrils; this process occurs often in structural proteins that consist of globular monomers that self-associate to form rigid fibers. Protein–protein interactions also regulate enzymatic activity, control progression through the cell cycle, and allow the assembly of large protein complexes that carry out many closely related reactions with a common biological function. Proteins can also bind to, or even be integrated into, cell membranes. The ability of binding partners to induce conformational changes in proteins allows the construction of enormously complex signaling
In signal processing, a signal is a function that conveys information about a phenomenon. Any quantity that can vary over space or time can be used as a signal to share messages between observers. The ''IEEE Transactions on Signal Processing'' ...
networks.
As interactions between proteins are reversible, and depend heavily on the availability of different groups of partner proteins to form aggregates that are capable to carry out discrete sets of function, study of the interactions between specific proteins is a key to understand important aspects of cellular function, and ultimately the properties that distinguish particular cell types.
Enzymes
The best-known role of proteins in the cell is as enzymes, which catalyse chemical reactions. Enzymes are usually highly specific and accelerate only one or a few chemical reactions. Enzymes carry out most of the reactions involved in metabolism, as well as manipulating DNA in processes such as DNA replication, DNA repair, andtranscription
Transcription refers to the process of converting sounds (voice, music etc.) into letters or musical notes, or producing a copy of something in another medium, including:
Genetics
* Transcription (biology), the copying of DNA into RNA, the fir ...
. Some enzymes act on other proteins to add or remove chemical groups in a process known as posttranslational modification. About 4,000 reactions are known to be catalysed by enzymes. The rate acceleration conferred by enzymatic catalysis is often enormous—as much as 1017-fold increase in rate over the uncatalysed reaction in the case of orotate decarboxylase
Orotidine 5'-phosphate decarboxylase (OMP decarboxylase) or orotidylate decarboxylase is an enzyme involved in pyrimidine biosynthesis. It catalyzes the decarboxylation of orotidine monophosphate (OMP) to form uridine monophosphate (UMP). The ...
(78 million years without the enzyme, 18 milliseconds with the enzyme).
The molecules bound and acted upon by enzymes are called substrates. Although enzymes can consist of hundreds of amino acids, it is usually only a small fraction of the residues that come in contact with the substrate, and an even smaller fraction—three to four residues on average—that are directly involved in catalysis. The region of the enzyme that binds the substrate and contains the catalytic residues is known as the active site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate (binding site) a ...
.
Dirigent proteins are members of a class of proteins that dictate the stereochemistry
Stereochemistry, a subdiscipline of chemistry, involves the study of the relative spatial arrangement of atoms that form the structure of molecules and their manipulation. The study of stereochemistry focuses on the relationships between stereois ...
of a compound synthesized by other enzymes.
Cell signaling and ligand binding
 Many proteins are involved in the process of cell signaling and
Many proteins are involved in the process of cell signaling and signal transduction
Signal transduction is the process by which a chemical or physical signal is transmitted through a cell as a series of molecular events, most commonly protein phosphorylation catalyzed by protein kinases, which ultimately results in a cellula ...
. Some proteins, such as insulin
Insulin (, from Latin ''insula'', 'island') is a peptide hormone produced by beta cells of the pancreatic islets encoded in humans by the ''INS'' gene. It is considered to be the main anabolic hormone of the body. It regulates the metabolism o ...
, are extracellular proteins that transmit a signal from the cell in which they were synthesized to other cells in distant tissues. Others are membrane proteins that act as receptors whose main function is to bind a signaling molecule and induce a biochemical response in the cell. Many receptors have a binding site exposed on the cell surface and an effector domain within the cell, which may have enzymatic activity or may undergo a conformational change
In biochemistry, a conformational change is a change in the shape of a macromolecule, often induced by environmental factors.
A macromolecule is usually flexible and dynamic. Its shape can change in response to changes in its environment or oth ...
detected by other proteins within the cell.
Antibodies
An antibody (Ab), also known as an immunoglobulin (Ig), is a large, Y-shaped protein used by the immune system to identify and neutralize foreign objects such as pathogenic bacteria and viruses. The antibody recognizes a unique molecule of the ...
are protein components of an adaptive immune system whose main function is to bind antigens, or foreign substances in the body, and target them for destruction. Antibodies can be secreted into the extracellular environment or anchored in the membranes of specialized B cell
B cells, also known as B lymphocytes, are a type of white blood cell of the lymphocyte subtype. They function in the humoral immunity component of the adaptive immune system. B cells produce antibody molecules which may be either secreted or ...
s known as plasma cell
Plasma cells, also called plasma B cells or effector B cells, are white blood cells that originate in the lymphoid organs as B lymphocytes and secrete large quantities of proteins called antibodies in response to being presented specific substan ...
s. Whereas enzymes are limited in their binding affinity for their substrates by the necessity of conducting their reaction, antibodies have no such constraints. An antibody's binding affinity to its target is extraordinarily high.
Many ligand transport proteins bind particular small biomolecules and transport them to other locations in the body of a multicellular organism. These proteins must have a high binding affinity when their ligand is present in high concentrations, but must also release the ligand when it is present at low concentrations in the target tissues. The canonical example of a ligand-binding protein is haemoglobin, which transports oxygen from the lung
The lungs are the primary organs of the respiratory system in humans and most other animals, including some snails and a small number of fish. In mammals and most other vertebrates, two lungs are located near the backbone on either side of t ...
s to other organs and tissues in all vertebrates and has close homologs in every biological kingdom. Lectins are sugar-binding proteins which are highly specific for their sugar moieties. Lectins typically play a role in biological recognition
Recognition may refer to:
*Award, something given in recognition of an achievement
Machine learning
*Pattern recognition, a branch of machine learning which encompasses the meanings below
Biometric
* Recognition of human individuals, or biomet ...
phenomena involving cells and proteins. Receptors and hormones are highly specific binding proteins.
Transmembrane proteins can also serve as ligand transport proteins that alter the permeability of the cell membrane to small molecules and ions. The membrane alone has a hydrophobic core through which polar
Polar may refer to:
Geography
Polar may refer to:
* Geographical pole, either of two fixed points on the surface of a rotating body or planet, at 90 degrees from the equator, based on the axis around which a body rotates
* Polar climate, the c ...
or charged molecules cannot diffuse. Membrane proteins contain internal channels that allow such molecules to enter and exit the cell. Many ion channel
Ion channels are pore-forming membrane proteins that allow ions to pass through the channel pore. Their functions include establishing a resting membrane potential, shaping action potentials and other electrical signals by gating the flow of io ...
proteins are specialized to select for only a particular ion; for example, potassium and sodium channels often discriminate for only one of the two ions.
Structural proteins
Structural proteins confer stiffness and rigidity to otherwise-fluid biological components. Most structural proteins are fibrous proteins; for example,collagen
Collagen () is the main structural protein in the extracellular matrix found in the body's various connective tissues. As the main component of connective tissue, it is the most abundant protein in mammals, making up from 25% to 35% of the whole ...
and elastin
Elastin is a protein that in humans is encoded by the ''ELN'' gene. Elastin is a key component of the extracellular matrix in gnathostomes (jawed vertebrates). It is highly elastic and present in connective tissue allowing many tissues in the bod ...
are critical components of connective tissue
Connective tissue is one of the four primary types of animal tissue, along with epithelial tissue, muscle tissue, and nervous tissue. It develops from the mesenchyme derived from the mesoderm the middle embryonic germ layer. Connective tiss ...
such as cartilage
Cartilage is a resilient and smooth type of connective tissue. In tetrapods, it covers and protects the ends of long bones at the joints as articular cartilage, and is a structural component of many body parts including the rib cage, the neck an ...
, and keratin is found in hard or filamentous structures such as hair
Hair is a protein filament that grows from follicles found in the dermis. Hair is one of the defining characteristics of mammals.
The human body, apart from areas of glabrous skin, is covered in follicles which produce thick terminal and f ...
, nails, feather
Feathers are epidermal growths that form a distinctive outer covering, or plumage, on both avian (bird) and some non-avian dinosaurs and other archosaurs. They are the most complex integumentary structures found in vertebrates and a premier ...
s, hooves, and some animal shell
An exoskeleton (from Greek ''éxō'' "outer" and ''skeletós'' "skeleton") is an external skeleton that supports and protects an animal's body, in contrast to an internal skeleton (endoskeleton) in for example, a human. In usage, some of the l ...
s. Some globular proteins can also play structural functions, for example, actin and tubulin are globular and soluble as monomers, but polymerize to form long, stiff fibers that make up the cytoskeleton, which allows the cell to maintain its shape and size.
Other proteins that serve structural functions are motor protein
Motor proteins are a class of molecular motors that can move along the cytoplasm of cells. They convert chemical energy into mechanical work by the hydrolysis of ATP. Flagellar rotation, however, is powered by a proton pump.
Cellular functions ...
s such as myosin
Myosins () are a superfamily of motor proteins best known for their roles in muscle contraction and in a wide range of other motility processes in eukaryotes. They are ATP-dependent and responsible for actin-based motility.
The first myosin ...
, kinesin, and dynein
Dyneins are a family of cytoskeletal motor proteins that move along microtubules in cells. They convert the chemical energy stored in ATP to mechanical work. Dynein transports various cellular cargos, provides forces and displacements importa ...
, which are capable of generating mechanical forces. These proteins are crucial for cellular motility of single celled organisms and the sperm
Sperm is the male reproductive cell, or gamete, in anisogamous forms of sexual reproduction (forms in which there is a larger, female reproductive cell and a smaller, male one). Animals produce motile sperm with a tail known as a flagellum, whi ...
of many multicellular organisms which reproduce sexually. They also generate the forces exerted by contracting muscle
Skeletal muscles (commonly referred to as muscles) are organs of the vertebrate muscular system and typically are attached by tendons to bones of a skeleton. The muscle cells of skeletal muscles are much longer than in the other types of muscl ...
s and play essential roles in intracellular transport.
Protein evolution
A key question in molecular biology is how proteins evolve, i.e. how can mutations (or rather changes in amino acid sequence) lead to new structures and functions? Most amino acids in a protein can be changed without disrupting activity or function, as can be seen from numeroushomologous
Homology may refer to:
Sciences
Biology
*Homology (biology), any characteristic of biological organisms that is derived from a common ancestor
*Sequence homology, biological homology between DNA, RNA, or protein sequences
* Homologous chrom ...
proteins across species (as collected in specialized databases for protein families
A protein family is a group of evolutionarily related proteins. In many cases, a protein family has a corresponding gene family, in which each gene encodes a corresponding protein with a 1:1 relationship. The term "protein family" should not be ...
, e.g. PFAM
Pfam is a database of protein families that includes their annotations and multiple sequence alignments generated using hidden Markov models. The most recent version, Pfam 35.0, was released in November 2021 and contains 19,632 families.
Uses
...
). In order to prevent dramatic consequences of mutations, a gene may be duplicated before it can mutate freely. However, this can also lead to complete loss of gene function and thus pseudo-genes. More commonly, single amino acid changes have limited consequences although some can change protein function substantially, especially in enzymes. For instance, many enzymes can change their substrate specificity by one or a few mutations. Changes in substrate specificity are facilitated by ''substrate promiscuity'', i.e. the ability of many enzymes to bind and process multiple substrates. When mutations occur, the specificity of an enzyme can increase (or decrease) and thus its enzymatic activity. Thus, bacteria (or other organisms) can adapt to different food sources, including unnatural substrates such as plastic.
Methods of study
The activities and structures of proteins may be examined '' in vitro,'' '' in vivo, andin silico
In biology and other experimental sciences, an ''in silico'' experiment is one performed on computer or via computer simulation. The phrase is pseudo-Latin for 'in silicon' (correct la, in silicio), referring to silicon in computer chips. It ...
''. ''In vitro'' studies of purified proteins in controlled environments are useful for learning how a protein carries out its function: for example, enzyme kinetics studies explore the chemical mechanism of an enzyme's catalytic activity and its relative affinity for various possible substrate molecules. By contrast, ''in vivo'' experiments can provide information about the physiological role of a protein in the context of a cell or even a whole organism. ''In silico'' studies use computational methods to study proteins.
Protein purification
To perform '' in vitro'' analysis, a protein must be purified away from other cellular components. This process usually begins with cell lysis, in which a cell's membrane is disrupted and its internal contents released into a solution known as acrude lysate
Lysis ( ) is the breaking down of the membrane of a cell, often by viral, enzymic, or osmotic (that is, "lytic" ) mechanisms that compromise its integrity. A fluid containing the contents of lysed cells is called a ''lysate''. In molecular bio ...
. The resulting mixture can be purified using ultracentrifugation, which fractionates the various cellular components into fractions containing soluble proteins; membrane lipids and proteins; cellular organelle
In cell biology, an organelle is a specialized subunit, usually within a cell, that has a specific function. The name ''organelle'' comes from the idea that these structures are parts of cells, as organs are to the body, hence ''organelle,'' the ...
s, and nucleic acid
Nucleic acids are biopolymers, macromolecules, essential to all known forms of life. They are composed of nucleotides, which are the monomers made of three components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main cl ...
s. Precipitation by a method known as salting out can concentrate the proteins from this lysate. Various types of chromatography are then used to isolate the protein or proteins of interest based on properties such as molecular weight, net charge and binding affinity. The level of purification can be monitored using various types of gel electrophoresis
Gel electrophoresis is a method for separation and analysis of biomacromolecules ( DNA, RNA, proteins, etc.) and their fragments, based on their size and charge. It is used in clinical chemistry to separate proteins by charge or size (IEF ...
if the desired protein's molecular weight and isoelectric point are known, by spectroscopy
Spectroscopy is the field of study that measures and interprets the electromagnetic spectra that result from the interaction between electromagnetic radiation and matter as a function of the wavelength or frequency of the radiation. Matter wa ...
if the protein has distinguishable spectroscopic features, or by enzyme assays if the protein has enzymatic activity. Additionally, proteins can be isolated according to their charge using electrofocusing
Isoelectric focusing (IEF), also known as electrofocusing, is a technique for separating different molecules by differences in their isoelectric point (pI). It is a type of zone electrophoresis usually performed on proteins in a gel that takes ad ...
.
For natural proteins, a series of purification steps may be necessary to obtain protein sufficiently pure for laboratory applications. To simplify this process, genetic engineering
Genetic engineering, also called genetic modification or genetic manipulation, is the modification and manipulation of an organism's genes using technology. It is a set of technologies used to change the genetic makeup of cells, including t ...
is often used to add chemical features to proteins that make them easier to purify without affecting their structure or activity. Here, a "tag" consisting of a specific amino acid sequence, often a series of histidine residues (a " His-tag"), is attached to one terminus of the protein. As a result, when the lysate is passed over a chromatography column containing nickel, the histidine residues ligate the nickel and attach to the column while the untagged components of the lysate pass unimpeded. A number of different tags have been developed to help researchers purify specific proteins from complex mixtures.
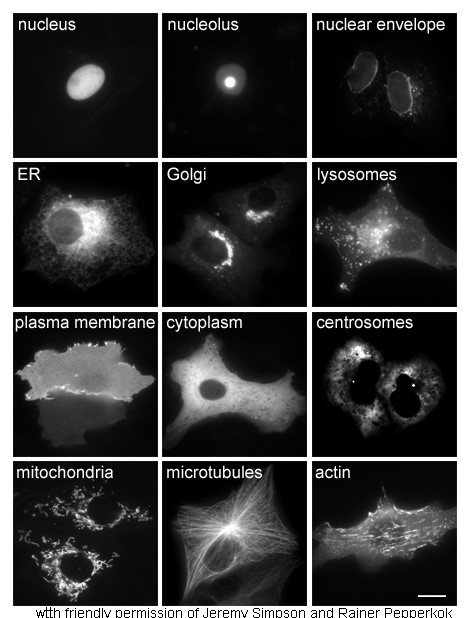
Cellular localization
 The study of proteins ''in vivo'' is often concerned with the synthesis and localization of the protein within the cell. Although many intracellular proteins are synthesized in the cytoplasm and membrane-bound or secreted proteins in the
The study of proteins ''in vivo'' is often concerned with the synthesis and localization of the protein within the cell. Although many intracellular proteins are synthesized in the cytoplasm and membrane-bound or secreted proteins in the endoplasmic reticulum
The endoplasmic reticulum (ER) is, in essence, the transportation system of the eukaryotic cell, and has many other important functions such as protein folding. It is a type of organelle made up of two subunits – rough endoplasmic reticulum ( ...
, the specifics of how proteins are targeted to specific organelles or cellular structures is often unclear. A useful technique for assessing cellular localization uses genetic engineering to express in a cell a fusion protein
Fusion proteins or chimeric (kī-ˈmir-ik) proteins (literally, made of parts from different sources) are proteins created through the joining of two or more genes that originally coded for separate proteins. Translation of this ''fusion gene'' r ...
or chimera consisting of the natural protein of interest linked to a "reporter
A journalist is an individual that collects/gathers information in form of text, audio, or pictures, processes them into a news-worthy form, and disseminates it to the public. The act or process mainly done by the journalist is called journalism ...
" such as green fluorescent protein
The green fluorescent protein (GFP) is a protein that exhibits bright green fluorescence when exposed to light in the blue to ultraviolet range. The label ''GFP'' traditionally refers to the protein first isolated from the jellyfish ''Aequorea ...
(GFP). The fused protein's position within the cell can be cleanly and efficiently visualized using microscopy
Microscopy is the technical field of using microscopes to view objects and areas of objects that cannot be seen with the naked eye (objects that are not within the resolution range of the normal eye). There are three well-known branches of micr ...
, as shown in the figure opposite.
Other methods for elucidating the cellular location of proteins requires the use of known compartmental markers for regions such as the ER, the Golgi, lysosomes or vacuoles, mitochondria, chloroplasts, plasma membrane, etc. With the use of fluorescently tagged versions of these markers or of antibodies to known markers, it becomes much simpler to identify the localization of a protein of interest. For example, indirect immunofluorescence
Immunofluorescence is a technique used for light microscopy with a fluorescence microscope and is used primarily on microbiological samples. This technique uses the specificity of antibodies to their antigen to target fluorescent dyes to spec ...
will allow for fluorescence colocalization and demonstration of location. Fluorescent dyes are used to label cellular compartments for a similar purpose.
Other possibilities exist, as well. For example, immunohistochemistry
Immunohistochemistry (IHC) is the most common application of immunostaining. It involves the process of selectively identifying antigens (proteins) in cells of a tissue section by exploiting the principle of antibodies binding specifically to an ...
usually uses an antibody to one or more proteins of interest that are conjugated to enzymes yielding either luminescent or chromogenic signals that can be compared between samples, allowing for localization information. Another applicable technique is cofractionation in sucrose (or other material) gradients using isopycnic centrifugation. While this technique does not prove colocalization of a compartment of known density and the protein of interest, it does increase the likelihood, and is more amenable to large-scale studies.
Finally, the gold-standard method of cellular localization is immunoelectron microscopy
An electron microscope is a microscope that uses a beam of accelerated electrons as a source of illumination. As the wavelength of an electron can be up to 100,000 times shorter than that of visible light photons, electron microscopes have a hi ...
. This technique also uses an antibody to the protein of interest, along with classical electron microscopy techniques. The sample is prepared for normal electron microscopic examination, and then treated with an antibody to the protein of interest that is conjugated to an extremely electro-dense material, usually gold. This allows for the localization of both ultrastructural details as well as the protein of interest.
Through another genetic engineering application known as site-directed mutagenesis, researchers can alter the protein sequence and hence its structure, cellular localization, and susceptibility to regulation. This technique even allows the incorporation of unnatural amino acids into proteins, using modified tRNAs, and may allow the rational design of new proteins with novel properties.
Proteomics
The total complement of proteins present at a time in a cell or cell type is known as its proteome, and the study of such large-scale data sets defines the field ofproteomics
Proteomics is the large-scale study of proteins. Proteins are vital parts of living organisms, with many functions such as the formation of structural fibers of muscle tissue, enzymatic digestion of food, or synthesis and replication of DNA. In ...
, named by analogy to the related field of genomics
Genomics is an interdisciplinary field of biology focusing on the structure, function, evolution, mapping, and editing of genomes. A genome is an organism's complete set of DNA, including all of its genes as well as its hierarchical, three-dim ...
. Key experimental techniques in proteomics include 2D electrophoresis, which allows the separation of many proteins, mass spectrometry
Mass spectrometry (MS) is an analytical technique that is used to measure the mass-to-charge ratio of ions. The results are presented as a ''mass spectrum'', a plot of intensity as a function of the mass-to-charge ratio. Mass spectrometry is use ...
, which allows rapid high-throughput identification of proteins and sequencing of peptides (most often after in-gel digestion), protein microarrays, which allow the detection of the relative levels of the various proteins present in a cell, and two-hybrid screening, which allows the systematic exploration of protein–protein interactions. The total complement of biologically possible such interactions is known as the interactome. A systematic attempt to determine the structures of proteins representing every possible fold is known as structural genomics.
Structure determination
Discovering the tertiary structure of a protein, or the quaternary structure of its complexes, can provide important clues about how the protein performs its function and how it can be affected, i.e. in drug design. As proteins are too small to be seen under alight microscope
The optical microscope, also referred to as a light microscope, is a type of microscope that commonly uses visible light and a system of lenses to generate magnified images of small objects. Optical microscopes are the oldest design of microsco ...
, other methods have to be employed to determine their structure. Common experimental methods include X-ray crystallography and NMR spectroscopy, both of which can produce structural information at atomic resolution. However, NMR experiments are able to provide information from which a subset of distances between pairs of atoms can be estimated, and the final possible conformations for a protein are determined by solving a distance geometry problem. Dual polarisation interferometry is a quantitative analytical method for measuring the overall protein conformation
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, the monomers of the polymer. A single amino acid monomer may ...
and conformational change
In biochemistry, a conformational change is a change in the shape of a macromolecule, often induced by environmental factors.
A macromolecule is usually flexible and dynamic. Its shape can change in response to changes in its environment or oth ...
s due to interactions or other stimulus. Circular dichroism is another laboratory technique for determining internal β-sheet / α-helical composition of proteins. Cryoelectron microscopy is used to produce lower-resolution structural information about very large protein complexes, including assembled viruses; a variant known as electron crystallography can also produce high-resolution information in some cases, especially for two-dimensional crystals of membrane proteins. Solved structures are usually deposited in the Protein Data Bank (PDB), a freely available resource from which structural data about thousands of proteins can be obtained in the form of Cartesian coordinates
A Cartesian coordinate system (, ) in a plane is a coordinate system that specifies each point uniquely by a pair of numerical coordinates, which are the signed distances to the point from two fixed perpendicular oriented lines, measured in t ...
for each atom in the protein.
Many more gene sequences are known than protein structures. Further, the set of solved structures is biased toward proteins that can be easily subjected to the conditions required in X-ray crystallography, one of the major structure determination methods. In particular, globular proteins are comparatively easy to crystallize in preparation for X-ray crystallography. Membrane proteins and large protein complexes, by contrast, are difficult to crystallize and are underrepresented in the PDB. Structural genomics initiatives have attempted to remedy these deficiencies by systematically solving representative structures of major fold classes. Protein structure prediction methods attempt to provide a means of generating a plausible structure for proteins whose structures have not been experimentally determined.
Structure prediction
 Complementary to the field of structural genomics, ''protein structure prediction'' develops efficient
Complementary to the field of structural genomics, ''protein structure prediction'' develops efficient mathematical model
A mathematical model is a description of a system using mathematical concepts and language. The process of developing a mathematical model is termed mathematical modeling. Mathematical models are used in the natural sciences (such as physics, ...
s of proteins to computationally predict the molecular formations in theory, instead of detecting structures with laboratory observation. The most successful type of structure prediction, known as homology modeling, relies on the existence of a "template" structure with sequence similarity to the protein being modeled; structural genomics' goal is to provide sufficient representation in solved structures to model most of those that remain. Although producing accurate models remains a challenge when only distantly related template structures are available, it has been suggested that sequence alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Alig ...
is the bottleneck in this process, as quite accurate models can be produced if a "perfect" sequence alignment is known. Many structure prediction methods have served to inform the emerging field of protein engineering
Protein engineering is the process of developing useful or valuable proteins. It is a young discipline, with much research taking place into the understanding of protein folding and recognition for protein design principles. It has been used to imp ...
, in which novel protein folds have already been designed. Also proteins (in eukaryotes ~33%) contain large unstructured but biologically functional segments and can be classified as intrinsically disordered proteins. Predicting and analysing protein disorder is, therefore, an important part of protein structure characterisation.
Bioinformatics
A vast array of computational methods have been developed to analyze the structure, function and evolution of proteins. The development of such tools has been driven by the large amount of genomic and proteomic data available for a variety of organisms, including the human genome. It is simply impossible to study all proteins experimentally, hence only a few are subjected to laboratory experiments while computational tools are used to extrapolate to similar proteins. Such homologous proteins can be efficiently identified in distantly related organisms bysequence alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Alig ...
. Genome and gene sequences can be searched by a variety of tools for certain properties. Sequence profiling tools can find restriction enzyme sites, open reading frames in nucleotide sequences, and predict secondary structure
Protein secondary structure is the three dimensional conformational isomerism, form of ''local segments'' of proteins. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta ...
s. Phylogenetic tree
A phylogenetic tree (also phylogeny or evolutionary tree Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA.) is a branching diagram or a tree showing the evolutionary relationships among various biological spec ...
s can be constructed and evolutionary hypotheses developed using special software like ClustalW
Clustal is a series of widely used computer programs used in bioinformatics for multiple sequence alignment. There have been many versions of Clustal over the development of the algorithm that are listed below. The analysis of each tool and its ...
regarding the ancestry of modern organisms and the genes they express. The field of bioinformatics
Bioinformatics () is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combi ...
is now indispensable for the analysis of genes and proteins.
In silico simulation of dynamical processes
A more complex computational problem is the prediction of intermolecular interactions, such as inmolecular docking
In the field of molecular modeling, docking is a method which predicts the preferred orientation of one molecule to a second when a ligand and a target are bound to each other to form a stable complex. Knowledge of the preferred orientation in tu ...
, protein folding, protein–protein interaction and chemical reactivity. Mathematical models to simulate these dynamical processes involve molecular mechanics, in particular, molecular dynamics. In this regard, ''in silico
In biology and other experimental sciences, an ''in silico'' experiment is one performed on computer or via computer simulation. The phrase is pseudo-Latin for 'in silicon' (correct la, in silicio), referring to silicon in computer chips. It ...
'' simulations discovered the folding of small α-helical protein domains such as the villin
Villin-1 is a 92.5 kDa tissue-specific actin-binding protein associated with the actin core bundle of the brush border. Villin-1 is encoded by the ''VIL1'' gene. Villin-1 contains multiple gelsolin-like domains capped by a small (8.5 kDa) "headp ...
headpiece, the HIV accessory protein and hybrid methods combining standard molecular dynamics with quantum mechanical mathematics have explored the electronic states of rhodopsin
Rhodopsin, also known as visual purple, is a protein encoded by the RHO gene and a G-protein-coupled receptor (GPCR). It is the opsin of the rod cells in the retina and a light-sensitive receptor protein that triggers visual phototransduction ...
s.
Beyond classical molecular dynamics, quantum dynamics methods allow the simulation of proteins in atomistic detail with an accurate description of quantum mechanical effects. Examples include the multi-layer multi-configuration time-dependent Hartree Multi-configuration time-dependent Hartree (MCTDH) is a general algorithm to solve the time-dependent Schrödinger equation
The Schrödinger equation is a linear partial differential equation that governs the wave function of a quantum-mecha ...
(MCTDH) method and the hierarchical equations of motion The hierarchical equations of motion (HEOM) technique derived by Yoshitaka Tanimura and Ryogo Kubo in 1989, is a non-perturbative approach developed to study the evolution of a density matrix \rho(t) of quantum dissipative systems. The method can t ...
(HEOM) approach, which have been applied to plant cryptochromes and bacteria light-harvesting complexes, respectively. Both quantum and classical mechanical simulations of biological-scale systems are extremely computationally demanding, so distributed computing initiatives (for example, the Folding@home project) facilitate the molecular modeling by exploiting advances in GPU parallel processing and Monte Carlo techniques.
Chemical analysis
The total nitrogen content of organic matter is mainly formed by the amino groups in proteins. The Total Kjeldahl Nitrogen (TKN
The Kjeldahl method or Kjeldahl digestion () in analytical chemistry is a method for the quantitative determination of nitrogen contained in organic substances plus the nitrogen contained in the inorganic compounds ammonia and ammonium (NH3/NH4+). ...
) is a measure of nitrogen widely used in the analysis of (waste) water, soil, food, feed and organic matter in general. As the name suggests, the Kjeldahl method
The Kjeldahl method or Kjeldahl digestion () in analytical chemistry is a method for the quantitative determination of nitrogen contained in organic substances plus the nitrogen contained in the inorganic compounds ammonia and ammonium (NH3/NH4+). ...
is applied. More sensitive methods are available.
Nutrition
Most microorganisms and plants can biosynthesize all 20 standardamino acids
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha am ...
, while animals (including humans) must obtain some of the amino acids from the diet. The amino acids that an organism cannot synthesize on its own are referred to as essential amino acids
An essential amino acid, or indispensable amino acid, is an amino acid that cannot be synthesized from scratch by the organism fast enough to supply its demand, and must therefore come from the diet. Of the 21 amino acids common to all life form ...
. Key enzymes that synthesize certain amino acids are not present in animals—such as aspartokinase
Aspartate kinase or aspartokinase (AK) is an enzyme that catalyzes the phosphorylation of the amino acid aspartate. This reaction is the first step in the biosynthesis of three other amino acids: methionine, lysine, and threonine, known as the " ...
, which catalyses the first step in the synthesis of lysine
Lysine (symbol Lys or K) is an α-amino acid that is a precursor to many proteins. It contains an α-amino group (which is in the protonated form under biological conditions), an α-carboxylic acid group (which is in the deprotonated −C ...
, methionine
Methionine (symbol Met or M) () is an essential amino acid in humans. As the precursor of other amino acids such as cysteine and taurine, versatile compounds such as SAM-e, and the important antioxidant glutathione, methionine plays a critical ro ...
, and threonine
Threonine (symbol Thr or T) is an amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form under biological conditions), a carboxyl group (which is in the deprotonated −COO� ...
from aspartate
Aspartic acid (symbol Asp or D; the ionic form is known as aspartate), is an α-amino acid that is used in the biosynthesis of proteins. Like all other amino acids, it contains an amino group and a carboxylic acid. Its α-amino group is in the pro ...
. If amino acids are present in the environment, microorganisms can conserve energy by taking up the amino acids from their surroundings and downregulating their biosynthetic pathways.
In animals, amino acids are obtained through the consumption of foods containing protein. Ingested proteins are then broken down into amino acids through digestion, which typically involves denaturation of the protein through exposure to acid
In computer science, ACID ( atomicity, consistency, isolation, durability) is a set of properties of database transactions intended to guarantee data validity despite errors, power failures, and other mishaps. In the context of databases, a sequ ...
and hydrolysis by enzymes called proteases. Some ingested amino acids are used for protein biosynthesis, while others are converted to glucose through gluconeogenesis
Gluconeogenesis (GNG) is a metabolic pathway that results in the generation of glucose from certain non-carbohydrate carbon substrates. It is a ubiquitous process, present in plants, animals, fungi, bacteria, and other microorganisms. In vertebrat ...
, or fed into the citric acid cycle. This use of protein as a fuel is particularly important under starvation
Starvation is a severe deficiency in caloric energy intake, below the level needed to maintain an organism's life. It is the most extreme form of malnutrition. In humans, prolonged starvation can cause permanent organ damage and eventually, dea ...
conditions as it allows the body's own proteins to be used to support life, particularly those found in muscle
Skeletal muscles (commonly referred to as muscles) are organs of the vertebrate muscular system and typically are attached by tendons to bones of a skeleton. The muscle cells of skeletal muscles are much longer than in the other types of muscl ...
.
In animals such as dogs and cats, protein maintains the health and quality of the skin by promoting hair follicle growth and keratinization, and thus reducing the likelihood of skin problems producing malodours. Poor-quality proteins also have a role regarding gastrointestinal health, increasing the potential for flatulence and odorous compounds in dogs because when proteins reach the colon in an undigested state, they are fermented producing hydrogen sulfide gas, indole, and skatole. Dogs and cats digest animal proteins better than those from plants, but products of low-quality animal origin are poorly digested, including skin, feathers, and connective tissue.
See also
References
Further reading
; Textbooks * * *External links
Databases and projects
NCBI Entrez Protein database
NCBI Protein Structure database
Human Protein Reference Database
Human Proteinpedia
Folding@Home (Stanford University)
Protein Databank in Europe
(see als
PDBeQuips
short articles and tutorials on interesting PDB structures)
Research Collaboratory for Structural Bioinformatics
(see als
, presenting short accounts on selected proteins from the PDB)
Proteopedia – Life in 3D
rotatable, zoomable 3D model with wiki annotations for every known protein molecular structure.
UniProt the Universal Protein Resource
Tutorials and educational websites
"An Introduction to Proteins"
from
HOPES
The Huntington's disease Outreach Project for Education at Stanford (HOPES) is a student-run project at Stanford University dedicated to making scientific information about Huntington's disease (HD) more readily accessible to patients and the publ ...
(Huntington's Disease Outreach Project for Education at Stanford)
Proteins: Biogenesis to Degradation – The Virtual Library of Biochemistry and Cell Biology
{{Authority control Molecular biology Proteomics