nonsense-mediated mRNA decay on:
[Wikipedia]
[Google]
[Amazon]
 Nonsense-mediated mRNA decay (NMD) is a surveillance pathway that exists in all
Nonsense-mediated mRNA decay (NMD) is a surveillance pathway that exists in all
doi: 10.1111/j.1365-2133.2012.11060
Published in the British Associations of Dermatologists in 2012, Lack of red hair phenotype in a North-African obese child homozygous for a novel POMC null mutation showed nonsense-mediated decay RNA evaluation in a hair pigment chemical analysis. They found that inactivating the POMC gene mutation results in obesity, adrenal insufficiency, and red hair. This has been seen in both in humans and mice. In this experiment they described a 3-year-old boy from Rome, Italy. He was a source of focus because he had
Yeast mRNA catabolism
Nonsense-Mediated Decay RNA Surveillance Pathway
Erroneous Gene Expression
{Dead link, date=April 2020 , bot=InternetArchiveBot , fix-attempted=yes
Decay in Health and Disease
Simplified Pathway Figure
* Holly, A.; Kuzmiak; Lynne E.M.; (2006) “Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges”. Department of Biochemistry and Biophysics. 12(7)306-316
doi:10.1016/j.molmed.2006.05.005
* Popp, M.W.; Maquat, L.E.; (2013) “Organizing principles of mammalian nonsense-mediated mRNA decay”. Annu Rev Genetics. 47:139-165
doi: 10.1146/annurev-genet-111212-133424
* Dang, Y.; Low, W.K.; (2009) “Inhibition of nonsense-mediated mRNA decay by the natural product pateamine A through eukaryotic initiation factor 4AIII”. J Biol Chem.284:23613–21
doi:10.1074/jbc.M109.009985
* Welch, E.M.; Barton, E.R.; (2007). “PTC124 targets genetic disorders caused by nonsense mutations”. Nature. 447:87–91
* Zou, T.; Mazan-Mamczarz, K.; (2006) “Polyamine depletion increases cytoplasmic levels of RNA-binding protein HuR leading to stabilization of nucleophosmin and p53 mRNAs”. J Biol Chem;281:19387–94
doi:10.1074/jbc.M602344200
* Zhang, Z.; Xin, D.; Wang, P.; (2009) “Noisy splicing, more than expression regulation, explains why some exons are subject to nonsense-mediated mRNA decay” . BMC Biol ;7:23
doi:10.1186/1741-7007-7-23
Gene expression
 Nonsense-mediated mRNA decay (NMD) is a surveillance pathway that exists in all
Nonsense-mediated mRNA decay (NMD) is a surveillance pathway that exists in all eukaryotes
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacte ...
. Its main function is to reduce errors in gene expression by eliminating mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein.
mRNA is ...
transcripts that contain premature stop codon
In molecular biology (specifically protein biosynthesis), a stop codon (or termination codon) is a codon (nucleotide triplet within messenger RNA) that signals the termination of the translation process of the current protein. Most codons in me ...
s. Translation of these aberrant mRNAs could, in some cases, lead to deleterious gain-of-function or dominant-negative activity of the resulting proteins.
NMD was first described in human cells and in yeast almost simultaneously in 1979. This suggested broad phylogenetic conservation and an important biological role of this intriguing mechanism. NMD was discovered when it was realized that cells often contain unexpectedly low concentrations of mRNAs that are transcribed from alleles carrying nonsense mutation
In genetics, a nonsense mutation is a point mutation in a sequence of DNA that results in a premature stop codon, or a ''nonsense codon'' in the transcribed mRNA, and in leading to a truncated, incomplete, and usually nonfunctional protein produc ...
s. Nonsense mutations code for a premature stop codon which causes the protein to be shortened. The truncated protein may or may not be functional, depending on the severity of what is not translated. In human genetics, NMD has the possibility to not only limit the translation of abnormal proteins, but it can occasionally cause detrimental effects in specific genetic mutations.
NMD functions to regulate numerous biological functions in a diverse range of cells, including the synaptic plasticity of neurons which may shape adult behavior.
Pathway
While many of the proteins involved in NMD are not conserved between species, in ''Saccharomyces cerevisiae'' (yeast), there are three main factors in NMD:UPF1
Regulator of nonsense transcripts 1 is a protein that in humans is encoded by the ''UPF1'' gene.
Function
This gene encodes a protein that is part of a post-splicing multiprotein complex, the exon junction complex, involved in both mRNA nuclea ...
, UPF2
Regulator of nonsense transcripts 2 is a protein that in humans is encoded by the ''UPF2'' gene.
Function
This gene encodes a protein that is part of a post-splicing multiprotein complex, the exon junction complex, involved in both mRNA nuclea ...
and UPF3 (UPF3A
Regulator of nonsense transcripts 3A is a protein that in humans is encoded by the ''UPF3A'' gene.
This gene encodes a protein that is part of a post-splicing multiprotein complex involved in both mRNA nuclear export and mRNA surveillance. The en ...
and UPF3B
Regulator of nonsense transcripts 3B is a protein that in humans is encoded by the ''UPF3B'' gene.
This gene encodes a protein that is part of a post-splicing multiprotein complex involved in both mRNA nuclear export and mRNA surveillance. The en ...
in humans), that make up the conserved core of the NMD pathway. All three of these factors are trans-acting In the field of molecular biology, ''trans''-acting (''trans''-regulatory, ''trans''-regulation), in general, means "acting from a different molecule" (''i.e.'', intermolecular). It may be considered the opposite of ''cis''-acting (''cis''-regulat ...
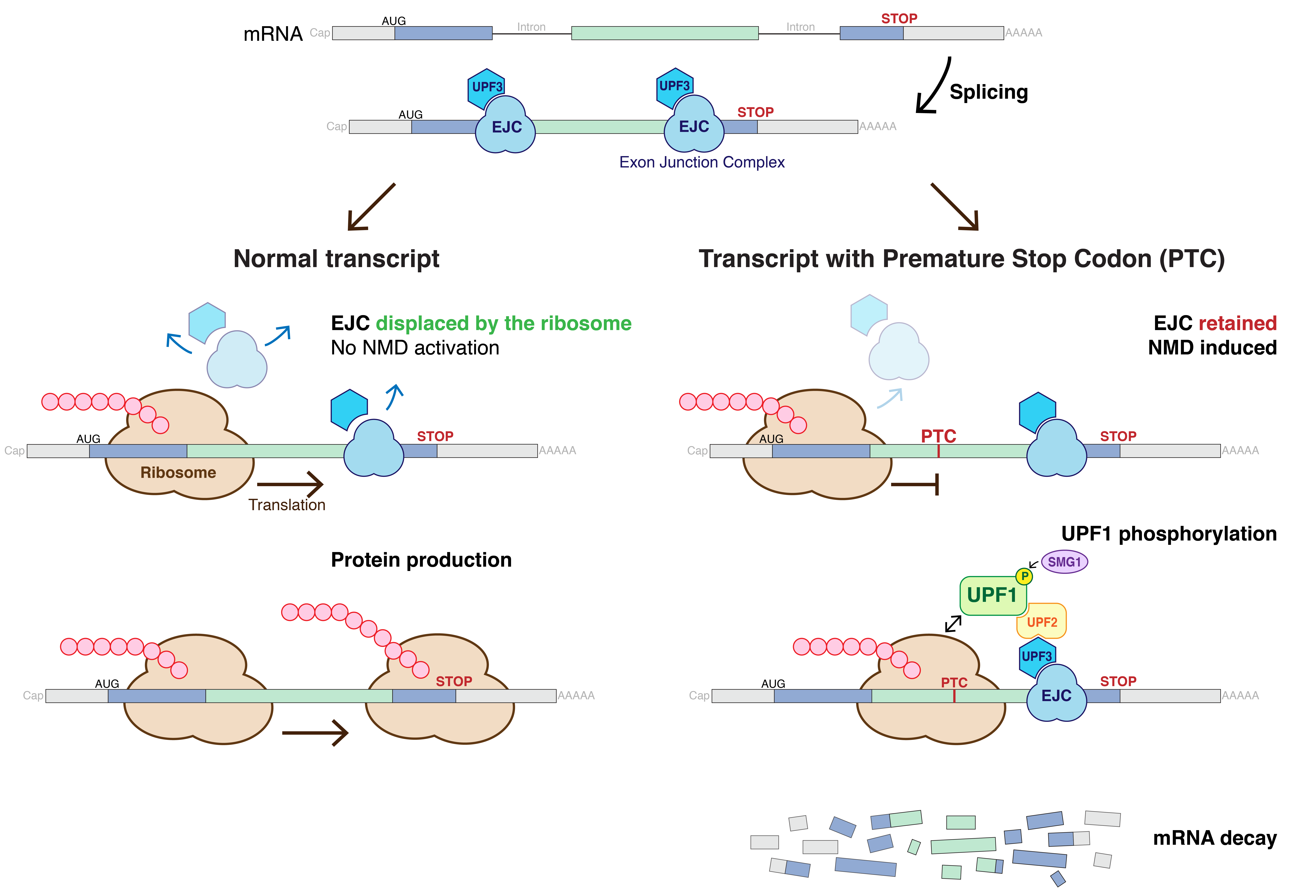
elements called up-frameshift (UPF) proteins. In mammals, UPF2 and UPF3 are part of the exon-exon junction complex (EJC) bound to mRNA after splicing along with other proteins, eIF4AIII, MLN51, and the Y14/MAGOH heterodimer, which also function in NMD. UPF1 phosphorylation is controlled by the proteins SMG-1, SMG-5, SMG-6 and SMG-7.
The process of detecting aberrant transcripts occurs during translation
Translation is the communication of the Meaning (linguistic), meaning of a #Source and target languages, source-language text by means of an Dynamic and formal equivalence, equivalent #Source and target languages, target-language text. The ...
of the mRNA. A popular model for the detection of aberrant transcripts in mammals suggests that during the first round of translation, the ribosome
Ribosomes ( ) are macromolecular machines, found within all cells, that perform biological protein synthesis (mRNA translation). Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to ...
removes the exon-exon junction complexes bound to the mRNA after splicing occurs. If after this first round of translation, any of these proteins remain bound to the mRNA, NMD is activated. Exon-exon junction complexes located downstream of a stop codon
In molecular biology (specifically protein biosynthesis), a stop codon (or termination codon) is a codon (nucleotide triplet within messenger RNA) that signals the termination of the translation process of the current protein. Most codons in me ...
are not removed from the transcript because the ribosome is released before reaching them. Termination of translation leads to the assembly of a complex composed of UPF1, SMG1 and the release factors, eRF1 and eRF3, on the mRNA. If an EJC is left on the mRNA because the transcript contains a premature stop codon, then UPF1 comes into contact with UPF2 and UPF3, triggering the phosphorylation
In chemistry, phosphorylation is the attachment of a phosphate group to a molecule or an ion. This process and its inverse, dephosphorylation, are common in biology and could be driven by natural selection. Text was copied from this source, wh ...
of UPF1. In vertebrates, the location of the last exon-junction complex relative to the termination codon usually determines whether the transcript will be subjected to NMD or not. If the termination codon is downstream of or within about 50 nucleotides of the final exon-junction complex then the transcript is translated normally. However, if the termination codon is further than about 50 nucleotides upstream of any exon-junction complexes, then the transcript is down regulated by NMD.Lewis BP, Green RE, Brenner SE. 2003. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proceedings of the National Academy of Sciences of the United States of America 100:189-192. doi:10.1016/j.bbrc.2009.04.021 The phosphorylated UPF1 then interacts with SMG-5, SMG-6 and SMG-7, which promote the dephosphorylation of UPF1. SMG-7 is thought to be the terminating effector in NMD, as it accumulates in P-bodies
P-bodies, or processing bodies are distinct foci formed by phase separation within the cytoplasm of the eukaryotic cell consisting of many enzymes involved in mRNA turnover. P-bodies are highly conserved structures and have been observed in soma ...
, which are cytoplasmic sites for mRNA decay. In both yeast and human cells, the major pathway for mRNA decay is initiated by the removal of the 5’ cap followed by degradation by XRN1, an exoribonuclease enzyme. The other pathway by which mRNA is degraded is by deadenylation
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In euka ...
from 3’-5'.
In addition to the well recognized role of NMD in removing aberrant transcripts, there are transcripts that contain introns within their 3'UTRs. These messages are predicted to be NMD-targets yet they (e.g., activity-regulated cytoskeleton-associated protein, known as Arc) can play crucial biologic functions suggesting that NMD may have physiologically relevant roles.
Mutations
Although nonsense-mediated mRNA decay reduces nonsense codons, mutations can occur that lead to various health problems and diseases in humans. A dominant-negative or deleterious gain-of-function mutation can occur if premature terminating (nonsense) codons are translated. NMD is becoming increasingly evident in the way it modifies phenotypic consequences because of the broad way it controls gene expression. For instance, the blood disorderBeta thalassemia
Beta thalassemias (β thalassemias) are a group of inherited blood disorders. They are forms of thalassemia caused by reduced or absent synthesis of the beta chains of hemoglobin that result in variable outcomes ranging from severe anemia to cli ...
is inherited and caused by mutations within the upstream of the β-globin gene. An individual carrying only one affected allele will have no or extremely low levels of the mutant β-globin mRNA. An even more severe form of the disease can occur called thalassemia intermedia or ‘inclusion body’ thalassemia. Instead of decreased mRNA levels, a mutant transcript produces truncated β chains, which in turn leads to a clinical phenotype in the heterozygote.
Nonsense-mediated decay mutations can also contribute to Marfan syndrome
Marfan syndrome (MFS) is a multi-systemic genetic disorder that affects the connective tissue. Those with the condition tend to be tall and thin, with long arms, legs, fingers, and toes. They also typically have exceptionally flexible joints a ...
. This disorder is caused by mutations in the fibrillin 1 (FBN1) gene and is resulted from a dominant negative interaction between mutant and wild-type fibrillin-1 gene.
Research applications
This pathway has a significant effect in the way genes are translated, restricting the amount of gene expression. It is still a new field in genetics, but its role in research has already led scientists to uncover numerous explanations for gene regulation. Studying nonsense-mediated decay has allowed scientists to determine the causes for certain heritable diseases and dosage compensation in mammals.Heritable diseases
Theproopiomelanocortin
Pro-opiomelanocortin (POMC) is a precursor polypeptide with 241 amino acid residues. POMC is synthesized in corticotrophs of the anterior pituitary from the 267-amino-acid-long polypeptide precursor pre-pro-opiomelanocortin (pre-POMC), by the r ...
gene (POMC) is expressed in the hypothalamus, in the pituitary gland. It yields a range of biologically active peptides and hormones and undergoes tissue-specific posttranslational processing to yield a range of biologically active peptides producing adrenocorticotropic hormone (ACTH), b-endorphin, and a-, b- and c-melanocyte-stimulating hormone (MSH). These peptides then interact with different melanocortin receptors (MCRs) and are involved in a wide range of processes including the regulation of body weight (MC3R and MC4R), adrenal steroidogenesis (MC2R) and hair pigmentation (MC1R).Cirillo, G.; Marini, R.; Ito, S.; (2012) “Lack of red hair phenotype in a North-African obese child homozygous for a novel POMC null mutation: nonsense-mediated decay RNA evaluation and hair pigment chemical analysis” . British Journal of Dermatology 167(6):1393-1395doi: 10.1111/j.1365-2133.2012.11060
Published in the British Associations of Dermatologists in 2012, Lack of red hair phenotype in a North-African obese child homozygous for a novel POMC null mutation showed nonsense-mediated decay RNA evaluation in a hair pigment chemical analysis. They found that inactivating the POMC gene mutation results in obesity, adrenal insufficiency, and red hair. This has been seen in both in humans and mice. In this experiment they described a 3-year-old boy from Rome, Italy. He was a source of focus because he had
Addison's disease
Addison's disease, also known as primary adrenal insufficiency, is a rare long-term endocrine disorder characterized by inadequate production of the steroid hormones cortisol and aldosterone by the two outer layers of the cells of the adrenal ...
and early onset obesity. They collected his DNA and amplified it using PCR. Sequencing analysis revealed a homozygous single substitution determining a stop codon. This caused an aberrant protein and the corresponding amino acid sequence indicated the exact position of the homozygous nucleotide. The substitution was localized in exon 3 and nonsense mutation at codon 68. The results from this experiment strongly suggest that the absence of red hair in non-European patients with early onset obesity and hormone deficiency does not exclude the occurrence of POMC mutations. By sequencing the patients DNA they found that this novel mutation has a collection of symptoms because of a malfunctioning nonsense-mediated mRNA decay surveillance pathway.
Dosage compensation
There has been evidence that the nonsense-mediated mRNA decay pathway participates in X chromosome dosage compensation in mammals. In higher eukaryotes with dimorphic sex chromosomes, such as humans and fruit flies, males have oneX chromosome
The X chromosome is one of the two sex-determining chromosomes (allosomes) in many organisms, including mammals (the other is the Y chromosome), and is found in both males and females. It is a part of the XY sex-determination system and XO sex-d ...
, whereas females have two. These organisms have evolved a mechanism that compensates not only for the different number of sex chromosomes between the two sexes, but also for the differing X/autosome ratios.Yin, S.;Deng, W.; Zheng, H.;(2009) “Evidence that the nonsense-mediated mRNA decay pathway participates in X chromosome dosage compensation in mammals”. Biochemical and Biophysical Research Communications 383(3)378–382. doi: 10.1016/j.bbrc.2009.04.021. In this genome-wide survey, the scientists found that autosomal genes are more likely to undergo nonsense-mediated decay than X-linked genes. This is because NMD fine tunes X chromosomes and it was demonstrated by inhibiting the pathway. The results were that balanced gene expression between X and autosomes gene expression was decreased by 10–15% no matter the method of inhibition. The NMD pathway is skewed towards depressing expression of larger population or autosomal genes than x-linked ones. In conclusion, the data supports the view that the coupling of alternative splicing and NMD is a pervasive means of gene expression regulation.
See also
*Non-stop decay
Non-stop decay (NSD) is a cellular mechanism of mRNA surveillance to detect mRNA molecules lacking a stop codon and prevent these mRNAs from translation. The non-stop decay pathway releases ribosomes that have reached the far 3' end of an mRNA and ...
, another mRNA surveillance mechanism
* mRNA surveillance
mRNA surveillance mechanisms are pathways utilized by organisms to ensure fidelity and quality of messenger RNA (mRNA) molecules. There are a number of surveillance mechanisms present within cells. These mechanisms function at various steps of th ...
References
External links
Yeast mRNA catabolism
Nonsense-Mediated Decay RNA Surveillance Pathway
Erroneous Gene Expression
{Dead link, date=April 2020 , bot=InternetArchiveBot , fix-attempted=yes
Decay in Health and Disease
Simplified Pathway Figure
* Holly, A.; Kuzmiak; Lynne E.M.; (2006) “Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges”. Department of Biochemistry and Biophysics. 12(7)306-316
doi:10.1016/j.molmed.2006.05.005
* Popp, M.W.; Maquat, L.E.; (2013) “Organizing principles of mammalian nonsense-mediated mRNA decay”. Annu Rev Genetics. 47:139-165
doi: 10.1146/annurev-genet-111212-133424
* Dang, Y.; Low, W.K.; (2009) “Inhibition of nonsense-mediated mRNA decay by the natural product pateamine A through eukaryotic initiation factor 4AIII”. J Biol Chem.284:23613–21
doi:10.1074/jbc.M109.009985
* Welch, E.M.; Barton, E.R.; (2007). “PTC124 targets genetic disorders caused by nonsense mutations”. Nature. 447:87–91
* Zou, T.; Mazan-Mamczarz, K.; (2006) “Polyamine depletion increases cytoplasmic levels of RNA-binding protein HuR leading to stabilization of nucleophosmin and p53 mRNAs”. J Biol Chem;281:19387–94
doi:10.1074/jbc.M602344200
* Zhang, Z.; Xin, D.; Wang, P.; (2009) “Noisy splicing, more than expression regulation, explains why some exons are subject to nonsense-mediated mRNA decay” . BMC Biol ;7:23
doi:10.1186/1741-7007-7-23
Gene expression