MicrobesOnline on:
[Wikipedia]
[Google]
[Amazon]
 MicrobesOnline is a publicly and freely accessible website that hosts multiple comparative genomic tools for comparing microbial species at the genomic, transcriptomic and functional levels. MicrobesOnline was developed by the Virtual Institute for Microbial Stress and Survival, which is based at the
MicrobesOnline is a publicly and freely accessible website that hosts multiple comparative genomic tools for comparing microbial species at the genomic, transcriptomic and functional levels. MicrobesOnline was developed by the Virtual Institute for Microbial Stress and Survival, which is based at the
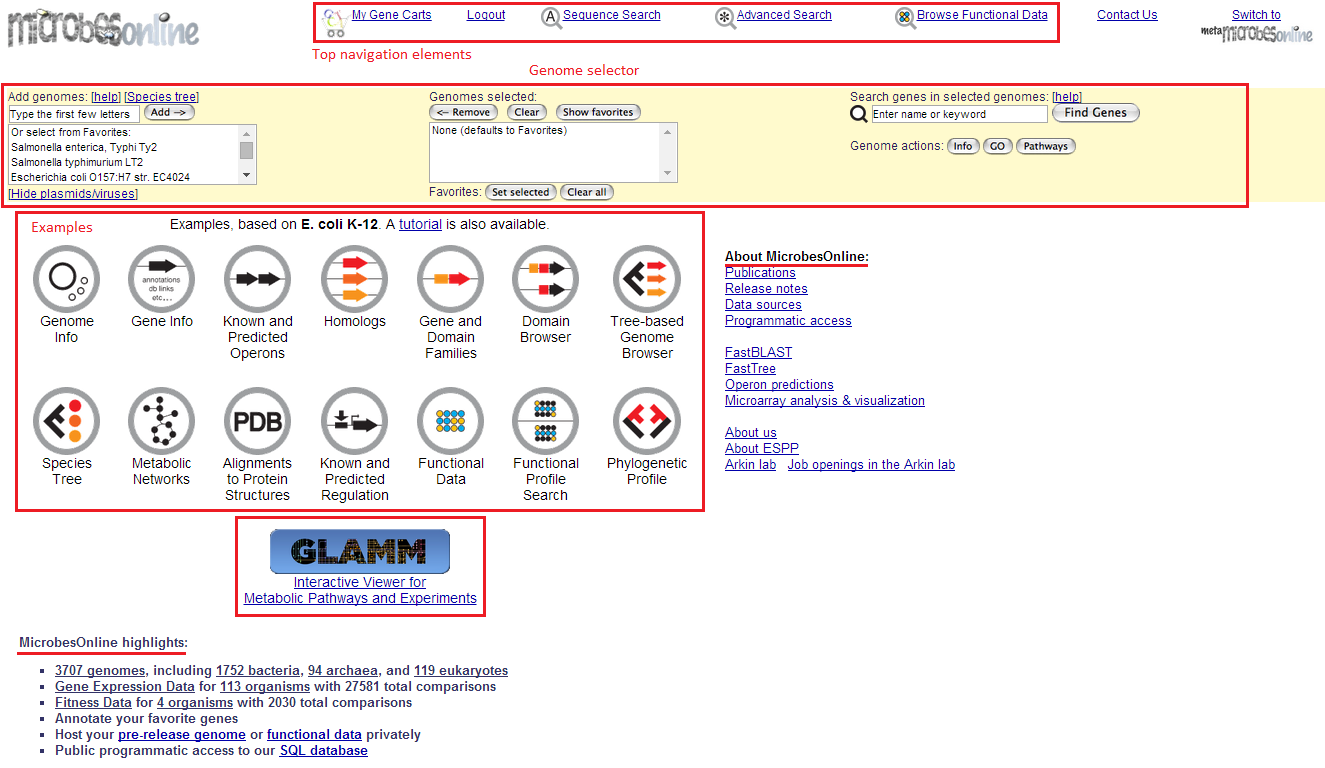 MicrobesOnline provides diverse tools for searching, analysing and integrating information related to bacteria genomes for applications in four major areas: genetic information, functional genomics, comparative genomics and metabolic pathway studies. The homepage of MicrobesOnline is the portal for accessing its functions, which includes six main sections: the top navigation elements, a genome selector, examples of the tutorial based on E.coli K-12, a link to the Genome-Linked Application for Metabolic Maps (GLAMM), website highlights and the “about MicrobesOnline” list. As an ongoing project, the authors of MicrobesOnline claim that the tools for data analysis and the support of more data types will be expanded.
MicrobesOnline provides diverse tools for searching, analysing and integrating information related to bacteria genomes for applications in four major areas: genetic information, functional genomics, comparative genomics and metabolic pathway studies. The homepage of MicrobesOnline is the portal for accessing its functions, which includes six main sections: the top navigation elements, a genome selector, examples of the tutorial based on E.coli K-12, a link to the Genome-Linked Application for Metabolic Maps (GLAMM), website highlights and the “about MicrobesOnline” list. As an ongoing project, the authors of MicrobesOnline claim that the tools for data analysis and the support of more data types will be expanded.
 The “genomes selected” box of the genome selector lists genomes added from the favourite genome list on the left or the ones searched by keywords. On the right side of the genome selector, four actions can be applied after selecting genomes: the “find genes” interface searches the gene name in the selected genomes and displays results in the gene list view; the “info” button lists a brief summary of selected genomes in the Summary View; the “GO” button opens a GO Browser called VertiGo which tabulates the number of genes under different GO items; finally, the “pathway” button initiates a pathway browser that illustrates the complete pathways of all organisms in the MicrobesOnline database.
In addition, the genome names shown in the summary view leads to a single-genome data view that presents a wealth of information about the selected genome. In the gene list view, the links “G O D H S T B...” lead the user to a locus information tool, where detailed information such as
The “genomes selected” box of the genome selector lists genomes added from the favourite genome list on the left or the ones searched by keywords. On the right side of the genome selector, four actions can be applied after selecting genomes: the “find genes” interface searches the gene name in the selected genomes and displays results in the gene list view; the “info” button lists a brief summary of selected genomes in the Summary View; the “GO” button opens a GO Browser called VertiGo which tabulates the number of genes under different GO items; finally, the “pathway” button initiates a pathway browser that illustrates the complete pathways of all organisms in the MicrobesOnline database.
In addition, the genome names shown in the summary view leads to a single-genome data view that presents a wealth of information about the selected genome. In the gene list view, the links “G O D H S T B...” lead the user to a locus information tool, where detailed information such as
 An important feature to store a user's work is the Gene Cart. Many web pages of MicrobesOnline displaying genetic information contain a link to add genes of interest to the session gene cart, which is available for all users. This is a temporary gene cart, and as such it loses information as a user closes the web browser. Genes in the session gene cart can be saved to the permanent gene cart which is only available to registered users after logging in.
An important feature to store a user's work is the Gene Cart. Many web pages of MicrobesOnline displaying genetic information contain a link to add genes of interest to the session gene cart, which is available for all users. This is a temporary gene cart, and as such it loses information as a user closes the web browser. Genes in the session gene cart can be saved to the permanent gene cart which is only available to registered users after logging in.
 The GO Browser, also known as VertiGo, is used by MicrobesOnline to search and visualise the GO hierarchy, which is a unified verbal system that describes properties of gene products, including cellular components, molecular function and biological process. The Genome Selector of the MicrobesOnline homepage provides a direct way to browse the GO hierarchy of the selected genomes, as well as provide a list of genes under a selected GO term, which can then be added to the session gene cart for further analysis.
The GO Browser, also known as VertiGo, is used by MicrobesOnline to search and visualise the GO hierarchy, which is a unified verbal system that describes properties of gene products, including cellular components, molecular function and biological process. The Genome Selector of the MicrobesOnline homepage provides a direct way to browse the GO hierarchy of the selected genomes, as well as provide a list of genes under a selected GO term, which can then be added to the session gene cart for further analysis.
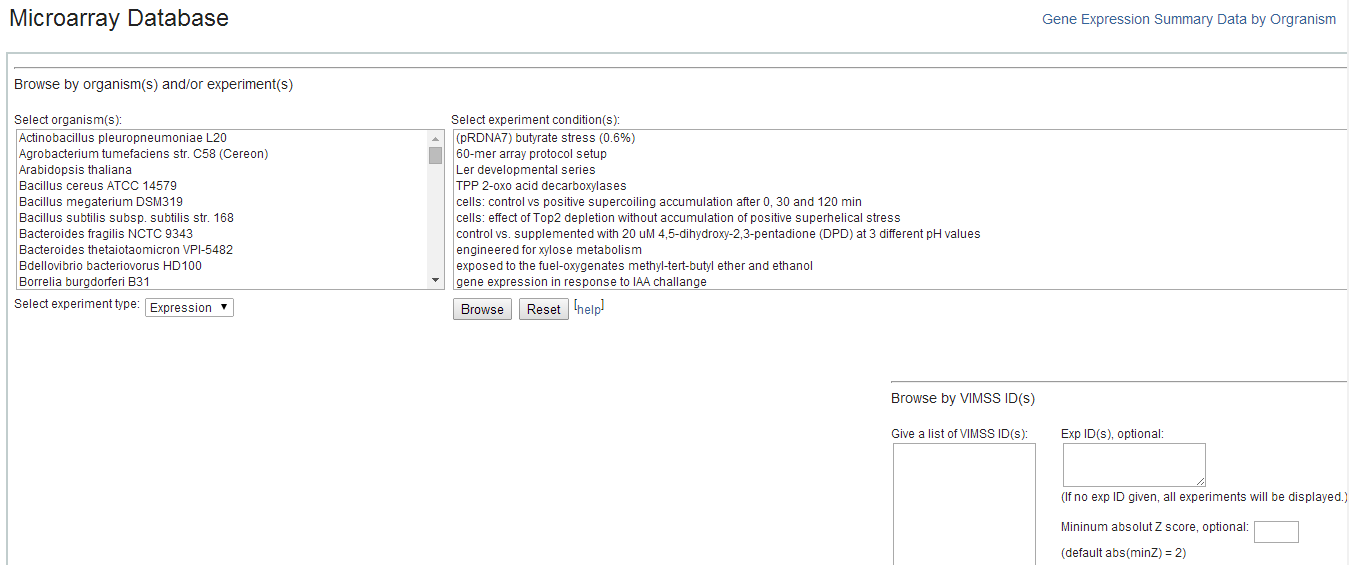 The Expression Data Viewer is an interface for searching and inspecting
The Expression Data Viewer is an interface for searching and inspecting
 The tree browser can be accessed by searching a gene by the Find Genes tool on the homepage with its VIMSS id (e.g. VIMSS15779). Once the gene context view has been accessed through the “Browse genomes by trees” option, a gene tree and a gene context diagram are displayed. In addition, the “View species tree” option opens a species tree view, which shows a species tree alongside the gene tree. Additionally, the tree browser enables users to choose both genes and genomes according to their similarity. Furthermore, it also demonstrates
The tree browser can be accessed by searching a gene by the Find Genes tool on the homepage with its VIMSS id (e.g. VIMSS15779). Once the gene context view has been accessed through the “Browse genomes by trees” option, a gene tree and a gene context diagram are displayed. In addition, the “View species tree” option opens a species tree view, which shows a species tree alongside the gene tree. Additionally, the tree browser enables users to choose both genes and genomes according to their similarity. Furthermore, it also demonstrates
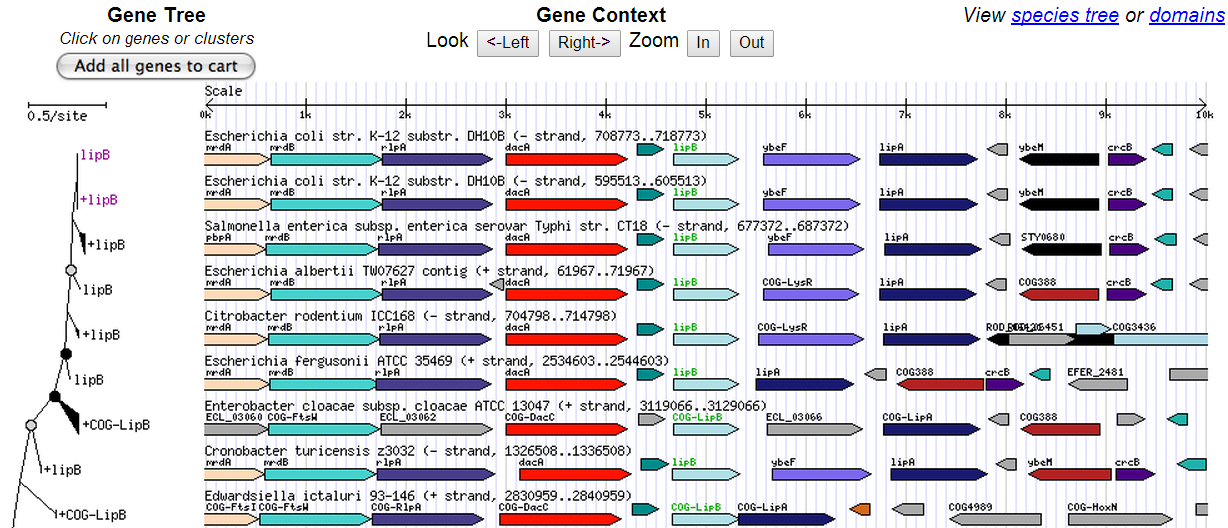 The Orthology Browser displays orthologs of genomes compared to the query genome by choosing multiple genomes from the “Select Organism(s) to Display” box.
The Orthology Browser displays orthologs of genomes compared to the query genome by choosing multiple genomes from the “Select Organism(s) to Display” box.  The locus information can be viewed through the “view genes” option, and this gene can be added to the session gene cart, or its gene expression data (including the heatmap) can be downloaded. Alternatively, a gene context view appears when browsing genomes by trees.
The locus information can be viewed through the “view genes” option, and this gene can be added to the session gene cart, or its gene expression data (including the heatmap) can be downloaded. Alternatively, a gene context view appears when browsing genomes by trees.
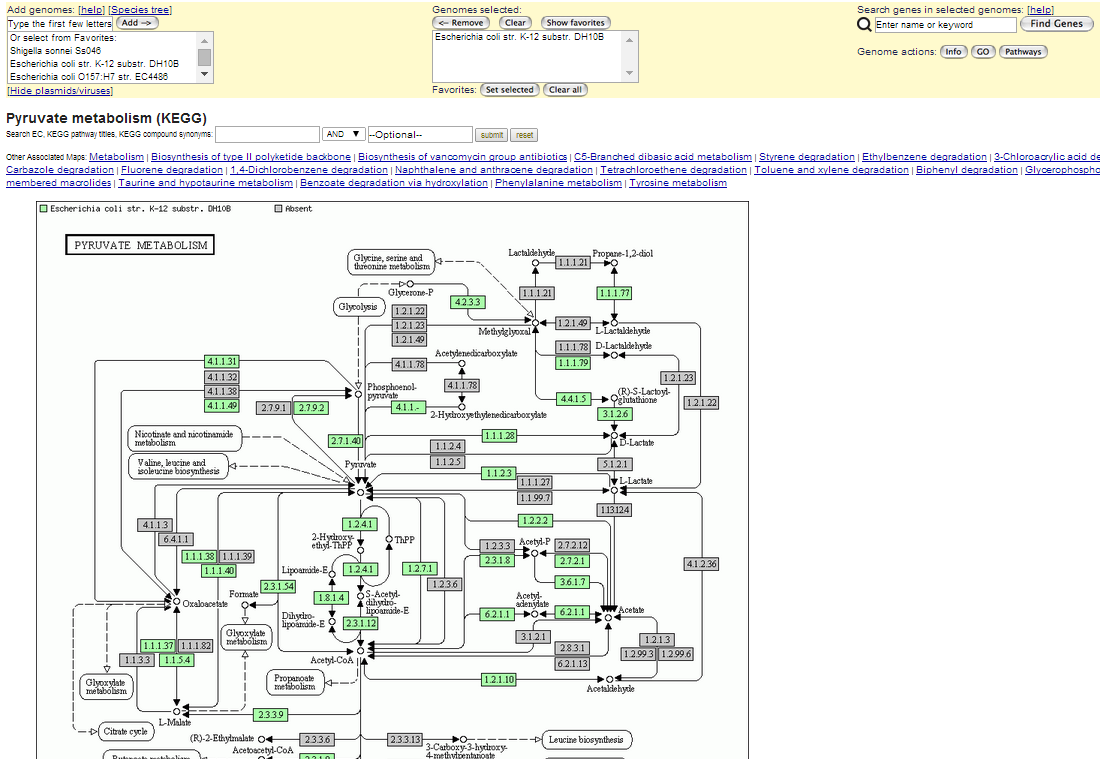
 The Pathway Browser lets users to navigate the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway maps displaying predicted presence or absence of enzymes for up to two selected genomes. The map of a particular pathway and a comparison between two kinds of microbes can be shown in the pathway browser. The enzyme commission number (e.g. 3.1.3.25) provides a link to the gene list view that shows information of the selected enzyme and allows the user to add genes to the session gene cart.
The Pathway Browser lets users to navigate the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway maps displaying predicted presence or absence of enzymes for up to two selected genomes. The map of a particular pathway and a comparison between two kinds of microbes can be shown in the pathway browser. The enzyme commission number (e.g. 3.1.3.25) provides a link to the gene list view that shows information of the selected enzyme and allows the user to add genes to the session gene cart.
 The GLAMM is another tool for searching and visualising metabolic pathways in a unified web interface. It helps users to identify or construct novel, transgenic pathways.
The GLAMM is another tool for searching and visualising metabolic pathways in a unified web interface. It helps users to identify or construct novel, transgenic pathways.
 MicrobesOnline is built on the integration of the data of an array of databases that manage different aspects of its capabilities. A comprehensive list is as follows:
*Sequence information: Non-redundant protein, gene and transcript sequences and annotations are extracted from
MicrobesOnline is built on the integration of the data of an array of databases that manage different aspects of its capabilities. A comprehensive list is as follows:
*Sequence information: Non-redundant protein, gene and transcript sequences and annotations are extracted from
MicrobesOnline home page
IMG home page
reference
Nucleic Acids Research, 2006, Vol. 34, Database issue D344-D348
Gene Ontology Consortium
(GOC): an international collaboration on developing gene ontology and creating annotations of genetic functions.
The Open Biological and Biomedical Ontologies
a community-based platform that integrates databases and tools of gene ontology.
Kyoto Encyclopedia of Genes and Genomes
(KEGG): a large collection of databases of genes, genomes, biological pathways, chemicals, drugs and diseases.
NCBI COGs
resources of phylogenetic studies of proteins encoded in complete genomes.
GLAMM
the Genome-Linked Application for Metabolic Maps, which is an interactive viewer for metabolic pathways and experiments.
a comprehensive guidance for using GLAMM.
NCBI's introduction to the MEGABLAST algorithm.
MultiExperiment Viewer: a versatile tool for analysing microarray data.
Microme
a platform that integrates a database, tools and browsers for studying bacterial metabolism.
MBGD
Microbial Genome Database for Comparative Analysis
Virtual Institute for Microbial Stress and Survival (VIMSS)
the supporter for an integrated program to study how microbes respond to and survive environmental stresses.
Lawrence Berkeley National Laboratory
Lawrence Berkeley National Laboratory (LBNL), commonly referred to as the Berkeley Lab, is a United States Department of Energy National Labs, United States national laboratory that is owned by, and conducts scientific research on behalf of, t ...
in Berkeley, California. The site was launched in 2005, with regular updates until 2011.
The main aim of MicrobesOnline is to provide an easy-to-use resource that integrates a wealth of data from multiple sources. This integrated platform facilitates studies in comparative genomics
Comparative genomics is a field of biological research in which the genomic features of different organisms are compared. The genomic features may include the DNA sequence, genes, gene order, regulatory sequences, and other genomic structural lan ...
, metabolic pathway
In biochemistry, a metabolic pathway is a linked series of chemical reactions occurring within a cell. The reactants, products, and intermediates of an enzymatic reaction are known as metabolites, which are modified by a sequence of chemical reac ...
analysis, genome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding ge ...
composition, functional genomics
Functional genomics is a field of molecular biology that attempts to describe gene (and protein) functions and interactions. Functional genomics make use of the vast data generated by genomic and transcriptomic projects (such as genome sequencing ...
as well as in protein domain
In molecular biology, a protein domain is a region of a protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist of s ...
and family
Family (from la, familia) is a Social group, group of people related either by consanguinity (by recognized birth) or Affinity (law), affinity (by marriage or other relationship). The purpose of the family is to maintain the well-being of its ...
data. It also provides tools to search or browse the database with genes, species, sequences, orthologous groups, gene ontology
The Gene Ontology (GO) is a major bioinformatics initiative to unify the representation of gene and gene product attributes across all species. More specifically, the project aims to: 1) maintain and develop its controlled vocabulary of gene and g ...
(GO) terms or pathway keywords, etc. Another one of its main features is the Gene Cart, which allows users to keep a record of their genes of interest. One of the highlights of the database is the overall navigation accessibility and interconnection between the tools.
Background
The development of high-throughput methods for genomesequencing
In genetics and biochemistry, sequencing means to determine the primary structure (sometimes incorrectly called the primary sequence) of an unbranched biopolymer. Sequencing results in a symbolic linear depiction known as a sequence which succ ...
has brought about a wealth of data that requires sophisticated bioinformatics
Bioinformatics () is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combi ...
tools for their analysis and interpretation. Nowadays, numerous tools exist to study genomics sequence data and extract information from different perspectives. However, the lack of unification of nomenclature and standardised protocols between tools, makes direct comparison between their results very difficult. Additionally, the user is forced to constantly switch from various websites or software, adjusting the format of their data to fit with individual requirements. MicrobesOnline was developed with the aim to integrate the capacities of different tools into a unified platform for easy comparison between analysis results, with a focus on prokaryote
A prokaryote () is a single-celled organism that lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Greek πρό (, 'before') and κάρυον (, 'nut' or 'kernel').Campbell, N. "Biology:Concepts & Connec ...
species and basal eukaryotes
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacte ...
.
Species included in the database
MicrobesOnline hosts genomic,gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. The ...
and fitness data for a wide range of microbial species. Genomic data is available for 1752 bacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among ...
, 94 archaea
Archaea ( ; singular archaeon ) is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria (in the Archaebac ...
and 119 eukaryotes, for a total of 3707 genomes, 2842 of which are marked as being complete. Gene expression data is available for 113 species, and fitness data is available for 4 organisms.
Functions and Site Architecture
 MicrobesOnline provides diverse tools for searching, analysing and integrating information related to bacteria genomes for applications in four major areas: genetic information, functional genomics, comparative genomics and metabolic pathway studies. The homepage of MicrobesOnline is the portal for accessing its functions, which includes six main sections: the top navigation elements, a genome selector, examples of the tutorial based on E.coli K-12, a link to the Genome-Linked Application for Metabolic Maps (GLAMM), website highlights and the “about MicrobesOnline” list. As an ongoing project, the authors of MicrobesOnline claim that the tools for data analysis and the support of more data types will be expanded.
MicrobesOnline provides diverse tools for searching, analysing and integrating information related to bacteria genomes for applications in four major areas: genetic information, functional genomics, comparative genomics and metabolic pathway studies. The homepage of MicrobesOnline is the portal for accessing its functions, which includes six main sections: the top navigation elements, a genome selector, examples of the tutorial based on E.coli K-12, a link to the Genome-Linked Application for Metabolic Maps (GLAMM), website highlights and the “about MicrobesOnline” list. As an ongoing project, the authors of MicrobesOnline claim that the tools for data analysis and the support of more data types will be expanded.
Genetic information
Information of microbial genes stored in MicrobesOnline includes sequences (gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a ba ...
s, transcripts and protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, respo ...
s), genomic loci, gene annotations and some statistics of sequences. This information can be accessed through three features displayed on the homepage of MicrobesOnline: sequence search and advanced search in the top navigation section, and the genome selector. For the sequence search tool, MicrobesOnline integrates BLAT, FastHMM and FastBLAST to search protein sequences, and uses MEGABLAST to search nucleotide sequences. It also provides a link to BLAST
Blast or The Blast may refer to:
* Explosion, a rapid increase in volume and release of energy in an extreme manner
*Detonation, an exothermic front accelerating through a medium that eventually drives a shock front
Film
* ''Blast'' (1997 film) ...
as an alternative way for searching sequences. On the other hand, the advanced search tool enables a user to search genetic information by categories, custom query, wild-card search and field-specific search, which uses the gene name, the description, the cluster of orthologous groups (COGs) id, the GO term, the KEGG
KEGG (Kyoto Encyclopedia of Genes and Genomes) is a collection of databases dealing with genomes, biological pathways, diseases, drugs, and chemical substances. KEGG is utilized for bioinformatics research and education, including data analysis i ...
enzyme commission (EC) number, etc. as key words.
 The “genomes selected” box of the genome selector lists genomes added from the favourite genome list on the left or the ones searched by keywords. On the right side of the genome selector, four actions can be applied after selecting genomes: the “find genes” interface searches the gene name in the selected genomes and displays results in the gene list view; the “info” button lists a brief summary of selected genomes in the Summary View; the “GO” button opens a GO Browser called VertiGo which tabulates the number of genes under different GO items; finally, the “pathway” button initiates a pathway browser that illustrates the complete pathways of all organisms in the MicrobesOnline database.
In addition, the genome names shown in the summary view leads to a single-genome data view that presents a wealth of information about the selected genome. In the gene list view, the links “G O D H S T B...” lead the user to a locus information tool, where detailed information such as
The “genomes selected” box of the genome selector lists genomes added from the favourite genome list on the left or the ones searched by keywords. On the right side of the genome selector, four actions can be applied after selecting genomes: the “find genes” interface searches the gene name in the selected genomes and displays results in the gene list view; the “info” button lists a brief summary of selected genomes in the Summary View; the “GO” button opens a GO Browser called VertiGo which tabulates the number of genes under different GO items; finally, the “pathway” button initiates a pathway browser that illustrates the complete pathways of all organisms in the MicrobesOnline database.
In addition, the genome names shown in the summary view leads to a single-genome data view that presents a wealth of information about the selected genome. In the gene list view, the links “G O D H S T B...” lead the user to a locus information tool, where detailed information such as operon
In genetics, an operon is a functioning unit of DNA containing a cluster of genes under the control of a single promoter. The genes are transcribed together into an mRNA strand and either translated together in the cytoplasm, or undergo splic ...
& regulon
In molecular genetics, a regulon is a group of genes that are regulated as a unit, generally controlled by the same regulatory gene that expresses a protein acting as a repressor or activator. This terminology is generally, although not exclusiv ...
, domains & families, sequences, annotations, etc. are shown.
Gene carts
 An important feature to store a user's work is the Gene Cart. Many web pages of MicrobesOnline displaying genetic information contain a link to add genes of interest to the session gene cart, which is available for all users. This is a temporary gene cart, and as such it loses information as a user closes the web browser. Genes in the session gene cart can be saved to the permanent gene cart which is only available to registered users after logging in.
An important feature to store a user's work is the Gene Cart. Many web pages of MicrobesOnline displaying genetic information contain a link to add genes of interest to the session gene cart, which is available for all users. This is a temporary gene cart, and as such it loses information as a user closes the web browser. Genes in the session gene cart can be saved to the permanent gene cart which is only available to registered users after logging in.
Functional genomics
One goal of setting up MicrobesOnline is to store functional information of microbial genomes. Such information includes gene ontology and microarray-based gene expression profiles, which can be accessed through two interfaces called GO browser and Expression Data Viewer respectively. The GO browser provides links to genes organised by gene ontology terms and the Expression Data Viewer provides both the access to expression profiles and information of experimental conditions.Gene ontology hierarchy
 The GO Browser, also known as VertiGo, is used by MicrobesOnline to search and visualise the GO hierarchy, which is a unified verbal system that describes properties of gene products, including cellular components, molecular function and biological process. The Genome Selector of the MicrobesOnline homepage provides a direct way to browse the GO hierarchy of the selected genomes, as well as provide a list of genes under a selected GO term, which can then be added to the session gene cart for further analysis.
The GO Browser, also known as VertiGo, is used by MicrobesOnline to search and visualise the GO hierarchy, which is a unified verbal system that describes properties of gene products, including cellular components, molecular function and biological process. The Genome Selector of the MicrobesOnline homepage provides a direct way to browse the GO hierarchy of the selected genomes, as well as provide a list of genes under a selected GO term, which can then be added to the session gene cart for further analysis.
Gene expression information
 The Expression Data Viewer is an interface for searching and inspecting
The Expression Data Viewer is an interface for searching and inspecting microarray
A microarray is a multiplex lab-on-a-chip. Its purpose is to simultaneously detect the expression of thousands of genes from a sample (e.g. from a tissue). It is a two-dimensional array on a solid substrate—usually a glass slide or silicon t ...
-base gene expression experiments and expression profiles. It consists of several components: an experiment browser for searching specific experiments in selected genomes under selected experimental conditions, an expression experiment viewer providing details of each microarray experiment, a gene expression viewer showing a heat map
A heat map (or heatmap) is a data visualization technique that shows magnitude of a phenomenon as color in two dimensions. The variation in color may be by hue or intensity, giving obvious visual cues to the reader about how the phenomenon is clu ...
of the expression levels of the selected gene and genes in the same operon
In genetics, an operon is a functioning unit of DNA containing a cluster of genes under the control of a single promoter. The genes are transcribed together into an mRNA strand and either translated together in the cytoplasm, or undergo splic ...
, and finally, a profile search tool for searching gene expression profiles. The Expression Data Viewer can be accessed through three ways: the “Browse Functional Data” in the navigator bar, the “Gene Expression Data” in the homepage and the “Gene expression” list in the single-genome data view, where the expression data are available. The single-genome data view can also show a protein-protein interaction browser that allows the inspection of interaction complexes and the download of expression data (e.g. Escherichia coli str. K-12 substr. MG1655). Furthermore, the user can launch a MultiExperiment Viewer (MeV) in the single-genome data view for analysing and visualising expression data.
Comparative genomics
MicrobesOnline stores information of genehomology
Homology may refer to:
Sciences
Biology
*Homology (biology), any characteristic of biological organisms that is derived from a common ancestor
* Sequence homology, biological homology between DNA, RNA, or protein sequences
*Homologous chrom ...
and phylogeny
A phylogenetic tree (also phylogeny or evolutionary tree Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA.) is a branching diagram or a tree showing the evolutionary relationships among various biological spec ...
for comparative genomic studies, which can be accessed through two interfaces. The first one is the Tree Browser, which draws a species tree or a gene tree for the selected gene and its gene neighbourhood. The second one is the Orthology Browser, which is an extension of the Genome Browser and demonstrates the selected gene within the context of its gene neighbourhood aligned with orthologs in other selected genomes. Both browsers provide options to save a gene in the session gene cart for further analysis.
Tree browser
 The tree browser can be accessed by searching a gene by the Find Genes tool on the homepage with its VIMSS id (e.g. VIMSS15779). Once the gene context view has been accessed through the “Browse genomes by trees” option, a gene tree and a gene context diagram are displayed. In addition, the “View species tree” option opens a species tree view, which shows a species tree alongside the gene tree. Additionally, the tree browser enables users to choose both genes and genomes according to their similarity. Furthermore, it also demonstrates
The tree browser can be accessed by searching a gene by the Find Genes tool on the homepage with its VIMSS id (e.g. VIMSS15779). Once the gene context view has been accessed through the “Browse genomes by trees” option, a gene tree and a gene context diagram are displayed. In addition, the “View species tree” option opens a species tree view, which shows a species tree alongside the gene tree. Additionally, the tree browser enables users to choose both genes and genomes according to their similarity. Furthermore, it also demonstrates horizontal gene transfer
Horizontal gene transfer (HGT) or lateral gene transfer (LGT) is the movement of genetic material between Unicellular organism, unicellular and/or multicellular organisms other than by the ("vertical") transmission of DNA from parent to offsprin ...
s among genomes.
Orthology browser
 The Orthology Browser displays orthologs of genomes compared to the query genome by choosing multiple genomes from the “Select Organism(s) to Display” box.
The Orthology Browser displays orthologs of genomes compared to the query genome by choosing multiple genomes from the “Select Organism(s) to Display” box.  The locus information can be viewed through the “view genes” option, and this gene can be added to the session gene cart, or its gene expression data (including the heatmap) can be downloaded. Alternatively, a gene context view appears when browsing genomes by trees.
The locus information can be viewed through the “view genes” option, and this gene can be added to the session gene cart, or its gene expression data (including the heatmap) can be downloaded. Alternatively, a gene context view appears when browsing genomes by trees.
Metabolic pathway information

 The Pathway Browser lets users to navigate the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway maps displaying predicted presence or absence of enzymes for up to two selected genomes. The map of a particular pathway and a comparison between two kinds of microbes can be shown in the pathway browser. The enzyme commission number (e.g. 3.1.3.25) provides a link to the gene list view that shows information of the selected enzyme and allows the user to add genes to the session gene cart.
The Pathway Browser lets users to navigate the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway maps displaying predicted presence or absence of enzymes for up to two selected genomes. The map of a particular pathway and a comparison between two kinds of microbes can be shown in the pathway browser. The enzyme commission number (e.g. 3.1.3.25) provides a link to the gene list view that shows information of the selected enzyme and allows the user to add genes to the session gene cart.
 The GLAMM is another tool for searching and visualising metabolic pathways in a unified web interface. It helps users to identify or construct novel, transgenic pathways.
The GLAMM is another tool for searching and visualising metabolic pathways in a unified web interface. It helps users to identify or construct novel, transgenic pathways.
Bioinformatics
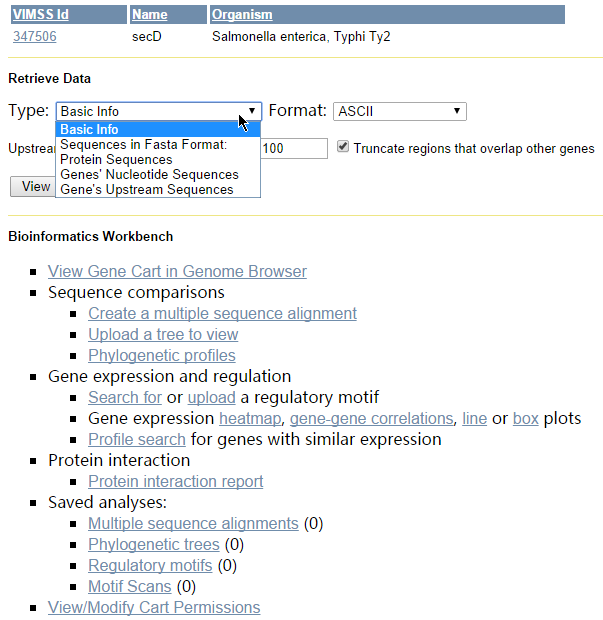
MicrobesOnline has integrated numerous tools for analysing sequences, gene expression profiles and protein-protein interactions into an interface called Bioinformatics Workbench, which is accessed via gene carts. Analyses currently supported includemultiple sequence alignment
Multiple sequence alignment (MSA) may refer to the process or the result of sequence alignment of three or more biological sequences, generally protein, DNA, or RNA. In many cases, the input set of query sequences are assumed to have an evolutio ...
s, construction of phylogenetic tree
A phylogenetic tree (also phylogeny or evolutionary tree Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA.) is a branching diagram or a tree showing the evolutionary relationships among various biological spec ...
s, motif searches and scans, summaries of gene expression profiles and protein-protein interactions. In order to save computational resources, a user is allowed to run two concurrent jobs for at most four hours and all results are saved temporarily until the session is terminated. Results can be shared with other users or groups via the resource access control tool.
Supporting databases
 MicrobesOnline is built on the integration of the data of an array of databases that manage different aspects of its capabilities. A comprehensive list is as follows:
*Sequence information: Non-redundant protein, gene and transcript sequences and annotations are extracted from
MicrobesOnline is built on the integration of the data of an array of databases that manage different aspects of its capabilities. A comprehensive list is as follows:
*Sequence information: Non-redundant protein, gene and transcript sequences and annotations are extracted from RefSeq
The Reference Sequence (RefSeq) database is an open access, annotated and curated collection of publicly available nucleotide sequences ( DNA, RNA) and their protein products. RefSeq was first introduced in 2000. This database is built by National ...
and Uniprot
UniProt is a freely accessible database of protein sequence and functional information, many entries being derived from genome sequencing projects. It contains a large amount of information about the biological function of proteins derived from ...
.
*Taxonomic classification of species and sequences: NCBI
The National Center for Biotechnology Information (NCBI) is part of the United States National Library of Medicine (NLM), a branch of the National Institutes of Health (NIH). It is approved and funded by the government of the United States. The ...
Taxonomy is used to classify the species and sequences into phylogenetic groups, and build a phylogenetic tree.
*Identification of non-annotated proteins from sequences: CRITICA is used to finds stretches of DNA sequence that code for proteins. Both a comparative genomics and a comparison and annotation-independent method are used.
*Identification of non-annotated genes from sequences: MicrobesOnline relies on Glimmer
In bioinformatics, GLIMMER (Gene Locator and Interpolated Markov ModelER) is used to find genes in prokaryotic DNA. "It is effective at finding genes in bacteria, archea, viruses, typically finding 98-99% of all relatively long protein coding g ...
to automatically find genes in bacteria, archaea and viral sequences.
*Classification of proteins: The classification of proteins by their conserved domain, family and superfamilies determined by PIRSF, Pfam
Pfam is a database of protein families that includes their annotations and multiple sequence alignments generated using hidden Markov models. The most recent version, Pfam 35.0, was released in November 2021 and contains 19,632 families.
Uses
...
, SMART
Smart or SMART may refer to:
Arts and entertainment
* ''Smart'' (Hey! Say! JUMP album), 2014
* Smart (Hotels.com), former mascot of Hotels.com
* ''Smart'' (Sleeper album), 1995 debut album by Sleeper
* ''SMart'', a children's television ser ...
and SUPERFAMILY
SUPERFAMILY is a database and search platform of structural and functional annotation for all proteins and genomes. It classifies amino acid sequences into known structural domains, especially into SCOP superfamilies. Domains are functional, str ...
repositories are included.
*Gene Orthology information: The orthologous groups of genes across species are based on the information on the COG database, which relies on protein sequence comparison for the detection of homology.
*Functional information of genes and proteins: The range of functional information provided is contributed by the following: GOA for Gene Ontology
The Gene Ontology (GO) is a major bioinformatics initiative to unify the representation of gene and gene product attributes across all species. More specifically, the project aims to: 1) maintain and develop its controlled vocabulary of gene and g ...
annotation of genes into functional categories, KEGG for metabolic, molecular and signaling pathways of genes, and PANTHER
Panther may refer to:
Large cats
*Pantherinae, the cat subfamily that contains the genera ''Panthera'' and ''Neofelis''
**''Panthera'', the cat genus that contains tigers, lions, jaguars and leopards.
***Jaguar (''Panthera onca''), found in Sout ...
for information about molecular and functional pathways, in the context of the relationships between protein families and their evolution . TIGRFAMs TIGRFAMs is a database of protein families designed to support manual and automated genome annotation. Each entry includes a multiple sequence alignment and hidden Markov model (HMM) built from the alignment. Sequences that score above the defined ...
and Gene3D are referred to for structural information and annotation of proteins.
*Gene expression data: Both NCBI GEO and Many Microbe Microarrays Database support the gene expression data of MicrobesOnline. The datasets compiled by Many Microbe Microarrays Database have the added advantage of being directly comparable, since only data generated by single-channel Affymetrix
Affymetrix is now Applied Biosystems, a brand of DNA microarray products sold by Thermo Fisher Scientific that originated with an American biotechnology research and development and manufacturing company of the same name. The Santa Clara, Califor ...
microarrays are accepted, and are subsequently normalised.
*Detection of CRISPRs: CRISPR
CRISPR () (an acronym for clustered regularly interspaced short palindromic repeats) is a family of DNA sequences found in the genomes of prokaryotic organisms such as bacteria and archaea. These sequences are derived from DNA fragments of bacte ...
are DNA loci involved in the immunity against invasive sequences, where short direct repeats are separated by spacer sequences. The databases generated by the CRT and PILER-CL algorithms are used to detect CRISPRs.
*Detection of tRNAs
Transfer RNA (abbreviated tRNA and formerly referred to as sRNA, for soluble RNA) is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes), that serves as the physical link between the mRNA and the amino a ...
: The tRNAscan-SE database is used as a reference to identify tRNA sequences.
*Submission of data by users: Users have the capacity of uploading both genomes and expression files to MicrobesOnline and analyse them with the analysis tools offered, with the option of keeping the data private (in the case of unpublished data) or releasing it to the public. Microarray data should include a clear identification of the organisms, platforms, treatments and controls, experimental conditions, time points and normalization techniques used, as well as the expression data in either log ratio or log levels format. Although draft genome sequences are accepted, they must be compliant with certain guidelines: (1) the assembled genome must have less than 100 scaffolds, (2) the FASTA
FASTA is a DNA and protein sequence alignment software package first described by David J. Lipman and William R. Pearson in 1985. Its legacy is the FASTA format which is now ubiquitous in bioinformatics.
History
The original FASTA program ...
file format should be used, having a unique label per contig
A contig (from ''contiguous'') is a set of overlapping DNA segments that together represent a consensus region of DNA.Gregory, S. ''Contig Assembly''. Encyclopedia of Life Sciences, 2005.
In bottom-up sequencing projects, a contig refers to ov ...
, (3) preferably gene predictions should be present (in this case, accepted formats include GenBank
The GenBank sequence database is an open access, annotated collection of all publicly available nucleotide sequences and their protein translations. It is produced and maintained by the National Center for Biotechnology Information (NCBI; a part ...
, EMBL
The European Molecular Biology Laboratory (EMBL) is an intergovernmental organization dedicated to molecular biology research and is supported by 27 member states, two prospect states, and one associate member state. EMBL was created in 1974 and ...
, tab-delimited and FASTA
FASTA is a DNA and protein sequence alignment software package first described by David J. Lipman and William R. Pearson in 1985. Its legacy is the FASTA format which is now ubiquitous in bioinformatics.
History
The original FASTA program ...
), (4) the name of the genome and the NCBI taxonomy ID should be provided.
Updates
MicrobesOnline was updated every 3 to 9 months from 2007 to 2011, where new features as well as new species data were added. However, there have been no new release notes since March 2011.Compatibility with other sites
MicrobesOnline is compatible with other similar platforms of integrated microbe data, such asIMG img or IMG is an abbreviation for image.
img or IMG may also refer to:
* IMG (company), global sports and media business headquartered in New York City but with its main offices in Cleveland, originally known as the "International Management Group ...
and RegTransBase
RegTransBase is database of regulatory interactions and transcription factor binding sites in prokaryotes
See also
*Transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein ...
, given that standard identifiers of genes are maintained throughout the database.
MicrobesOnline in the realm of microbe analysis platforms
There have been other efforts to create a unified platform for prokaryote analysis tools, however, most of them focus on one set of analysis types. A few examples of these focused databases include those with an emphasis on metabolic data analysis (Microme ), comparative genomics (MBGD and the OMA Browser ),regulon
In molecular genetics, a regulon is a group of genes that are regulated as a unit, generally controlled by the same regulatory gene that expresses a protein acting as a repressor or activator. This terminology is generally, although not exclusiv ...
s and transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The fu ...
s (RegPrecise ), comparative functional genomics (Pathline ), among many others. However, notable efforts have been made by other teams to create comprehensive platforms that largely overlap with the capabilities of MicrobesOnline. MicroScope
A microscope () is a laboratory instrument used to examine objects that are too small to be seen by the naked eye. Microscopy is the science of investigating small objects and structures using a microscope. Microscopic means being invisibl ...
and the Integrated Microbial Genomes System
The Integrated Microbial GenomesIMG system is a genome browsing and annotation platform developed by the U.S. Department of Energy (DOE)-Joint Genome Institute. IMG contains all the draft and complete microbial genomes sequenced by the DOE-JGI in ...
(IMG) are examples of popular and recently updated databases ().
Extension of metagenome analysis: metaMicrobesOnline
metaMicrobesOnline was compiled by the same developers as MicrobesOnline, and constitutes an extension of MicrobesOnline capacities, by focusing on the phylogenetic analysis of metagenomes. With a similar web interface to MicrobesOnline, the user is capable of toggling between sites via the “switch to” link on the homepage.See also
*Integrated Microbial Genomes System
The Integrated Microbial GenomesIMG system is a genome browsing and annotation platform developed by the U.S. Department of Energy (DOE)-Joint Genome Institute. IMG contains all the draft and complete microbial genomes sequenced by the DOE-JGI in ...
(IMG)
*BLAST
Blast or The Blast may refer to:
* Explosion, a rapid increase in volume and release of energy in an extreme manner
*Detonation, an exothermic front accelerating through a medium that eventually drives a shock front
Film
* ''Blast'' (1997 film) ...
*BLAT (bioinformatics)
BLAT (BLAST-like alignment tool) is a pairwise sequence alignment algorithm that was developed by Jim Kent at the University of California Santa Cruz (UCSC) in the early 2000s to assist in the assembly and annotation of the human genome. It was de ...
*FASTA format
In bioinformatics and biochemistry, the FASTA format is a text-based format for representing either nucleotide sequences or amino acid (protein) sequences, in which nucleotides or amino acids are represented using single-letter codes. The format a ...
*Gene ontology
The Gene Ontology (GO) is a major bioinformatics initiative to unify the representation of gene and gene product attributes across all species. More specifically, the project aims to: 1) maintain and develop its controlled vocabulary of gene and g ...
* Hidden Markov model (HMM)
*Homology (biology)
In biology, homology is similarity due to shared ancestry between a pair of structures or genes in different taxa. A common example of homologous structures is the forelimbs of vertebrates, where the wings of bats and birds, the arms of pri ...
*KEGG
KEGG (Kyoto Encyclopedia of Genes and Genomes) is a collection of databases dealing with genomes, biological pathways, diseases, drugs, and chemical substances. KEGG is utilized for bioinformatics research and education, including data analysis i ...
*Multiple Sequence Alignment
Multiple sequence alignment (MSA) may refer to the process or the result of sequence alignment of three or more biological sequences, generally protein, DNA, or RNA. In many cases, the input set of query sequences are assumed to have an evolutio ...
External links
MicrobesOnline home page
IMG home page
reference
Nucleic Acids Research, 2006, Vol. 34, Database issue D344-D348
Gene Ontology Consortium
(GOC): an international collaboration on developing gene ontology and creating annotations of genetic functions.
The Open Biological and Biomedical Ontologies
a community-based platform that integrates databases and tools of gene ontology.
Kyoto Encyclopedia of Genes and Genomes
(KEGG): a large collection of databases of genes, genomes, biological pathways, chemicals, drugs and diseases.
NCBI COGs
resources of phylogenetic studies of proteins encoded in complete genomes.
GLAMM
the Genome-Linked Application for Metabolic Maps, which is an interactive viewer for metabolic pathways and experiments.
a comprehensive guidance for using GLAMM.
NCBI's introduction to the MEGABLAST algorithm.
MultiExperiment Viewer: a versatile tool for analysing microarray data.
Microme
a platform that integrates a database, tools and browsers for studying bacterial metabolism.
MBGD
Microbial Genome Database for Comparative Analysis
Virtual Institute for Microbial Stress and Survival (VIMSS)
the supporter for an integrated program to study how microbes respond to and survive environmental stresses.
References
{{genomics-footer * Genome databases