|
Integrated Microbial Genomes System
The Integrated Microbial GenomesIMG system is a genome browsing and annotation platform developed by the U.S. Department of Energy (DOE)-Joint Genome Institute. IMG contains all the draft and complete microbial genomes sequenced by the DOE-JGI integrated with other publicly available genomes (including Archaea, Bacteria, Eukarya, Viruses and Plasmids). IMG provides users a set of tools for comparative analysis of microbial genomes along three dimensions: genes, genomes and functions. Users can select and transfer them in the comparative analysis carts based upon a variety of criteria. IMG also includes a genome annotation pipeline that integrates information from several tools, including KEGG, Pfam, InterPro, and the Gene Ontology, among others. Users can also type or upload their own gene annotations (called MyIMG gene annotations) and the IMG system will allow them to generate Genbank or EMBL format files containing these annotations. In successive releases IMG has expanded to i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
United States Department Of Energy
The United States Department of Energy (DOE) is an executive department of the U.S. federal government that oversees U.S. national energy policy and manages the research and development of nuclear power and nuclear weapons in the United States. The DOE oversees the U.S. nuclear weapons program, nuclear reactor production for the United States Navy, energy-related research, and domestic energy production and energy conservation. The DOE was created in 1977 in the aftermath of the 1973 oil crisis. It sponsors more physical science research than any other U.S. federal agency, the majority of which is conducted through its system of National Laboratories. The DOE also directs research in genomics, with the Human Genome Project originating from a DOE initiative. The department is headed by the Secretary of Energy, who reports directly to the president of the United States and is a member of the Cabinet. The current Secretary of Energy is Jennifer Granholm, who has served ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Joint Genome Institute
The U.S. Department of Energy (DOE) Joint Genome Institute (JGI), first located in Walnut Creek then Berkeley, California, was created in 1997 to unite the expertise and resources in genome mapping, DNA sequencing, technology development, and information sciences pioneered at the DOE genome centers at Lawrence Berkeley National Laboratory (Berkeley Lab), Lawrence Livermore National Laboratory (LLNL) and Los Alamos National Laboratory (LANL). As a DOE Office of Science User Facility of Berkeley Lab, the JGI staff is composed of employees from Berkeley Lab, LLNL and the HudsonAlpha Institute for Biotechnology. The JGI also collaborates with other DOE-supported programs and facilities, such as the Environmental Molecular Sciences Laboratory at Pacific Northwest National Laboratory (PNNL), the National Energy Research Scientific Computing Center, or NERSC, and the DOE Bioenergy Research Centers. History In 1999, the University of California, which manages the three national labs for ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
KEGG
KEGG (Kyoto Encyclopedia of Genes and Genomes) is a collection of databases dealing with genomes, biological pathways, diseases, drugs, and chemical substances. KEGG is utilized for bioinformatics research and education, including data analysis in genomics, metagenomics, metabolomics and other omics studies, modeling and simulation in systems biology, and translational research in drug development. The KEGG database project was initiated in 1995 by Minoru Kanehisa, professor at the Institute for Chemical Research, Kyoto University, under the then ongoing Japanese Human Genome Program. Foreseeing the need for a computerized resource that can be used for biological interpretation of genome sequence data, he started developing the KEGG PATHWAY database. It is a collection of manually drawn KEGG pathway maps representing experimental knowledge on metabolism and various other functions of the cell and the organism. Each pathway map contains a network of molecular interactions and re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pfam
Pfam is a database of protein families that includes their annotations and multiple sequence alignments generated using hidden Markov models. The most recent version, Pfam 35.0, was released in November 2021 and contains 19,632 families. Uses The general purpose of the Pfam database is to provide a complete and accurate classification of protein families and domains. Originally, the rationale behind creating the database was to have a semi-automated method of curating information on known protein families to improve the efficiency of annotating genomes. The Pfam classification of protein families has been widely adopted by biologists because of its wide coverage of proteins and sensible naming conventions. It is used by experimental biologists researching specific proteins, by structural biologists to identify new targets for structure determination, by computational biologists to organise sequences and by evolutionary biologists tracing the origins of proteins. Early genome ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
InterPro
InterPro is a database of protein families, protein domains and functional sites in which identifiable features found in known proteins can be applied to new protein sequences in order to functionally characterise them. The contents of InterPro consist of diagnostic signatures and the proteins that they significantly match. The signatures consist of models (simple types, such as regular expressions or more complex ones, such as Hidden Markov models) which describe protein families, domains or sites. Models are built from the amino acid sequences of known families or domains and they are subsequently used to search unknown sequences (such as those arising from novel genome sequencing) in order to classify them. Each of the member databases of InterPro contributes towards a different niche, from very high-level, structure-based classifications ( SUPERFAMILY and CATH-Gene3D) through to quite specific sub-family classifications ( PRINTS and PANTHER). InterPro's intention is to pro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Ontology
The Gene Ontology (GO) is a major bioinformatics initiative to unify the representation of gene and gene product attributes across all species. More specifically, the project aims to: 1) maintain and develop its controlled vocabulary of gene and gene product attributes; 2) annotate genes and gene products, and assimilate and disseminate annotation data; and 3) provide tools for easy access to all aspects of the data provided by the project, and to enable functional interpretation of experimental data using the GO, for example via enrichment analysis. GO is part of a larger classification effort, the Open Biomedical Ontologies, being one of the Initial Candidate Members of the OBO Foundry. Whereas gene nomenclature focuses on gene and gene products, the Gene Ontology focuses on the function of the genes and gene products. The GO also extends the effort by using markup language to make the data (not only of the genes and their products but also of curated attributes) machine read ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genbank
The GenBank sequence database is an open access, annotated collection of all publicly available nucleotide sequences and their protein translations. It is produced and maintained by the National Center for Biotechnology Information (NCBI; a part of the National Institutes of Health in the United States) as part of the International Nucleotide Sequence Database Collaboration (INSDC). GenBank and its collaborators receive sequences produced in laboratories throughout the world from more than 500,000 formally described species. The database started in 1982 by Walter Goad and Los Alamos National Laboratory. GenBank has become an important database for research in biological fields and has grown in recent years at an exponential rate by doubling roughly every 18 months. Release 250.0, published in June 2022, contained over 17 trillion nucleotide bases in more than 2,45 billion sequences. GenBank is built by direct submissions from individual laboratories, as well as from bulk submis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EMBL
The European Molecular Biology Laboratory (EMBL) is an intergovernmental organization dedicated to molecular biology research and is supported by 27 member states, two prospect states, and one associate member state. EMBL was created in 1974 and is funded by public research money from its member states. Research at EMBL is conducted by approximately 110 independent research and service groups and teams covering the spectrum of molecular biology and bioinformatics. The list of Groups and Teams at EMBL can be found at . The Laboratory operates from six sites: the main laboratory in Heidelberg, and sites in Hinxton (the European Bioinformatics Institute (EBI), in England), Grenoble (France), Hamburg (Germany), Rome (Italy) and Barcelona (Spain). EMBL groups and laboratories perform basic research in molecular biology and molecular medicine as well as train scientists, students, and visitors. The organization aids in the development of services, new instruments and methods, and techno ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
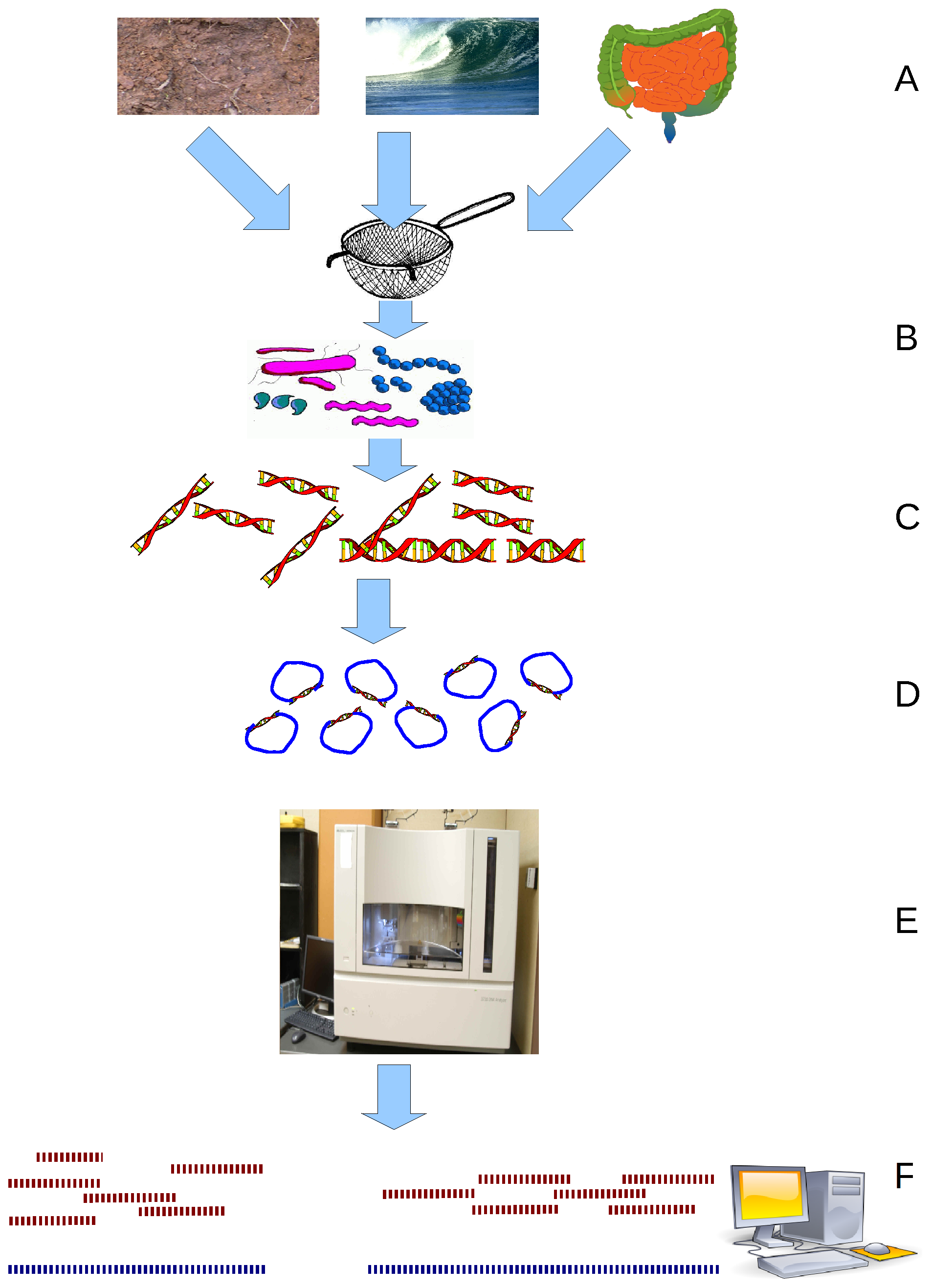
Metagenomics
Metagenomics is the study of genetic material recovered directly from environmental or clinical samples by a method called sequencing. The broad field may also be referred to as environmental genomics, ecogenomics, community genomics or microbiomics. While traditional microbiology and microbial genome sequencing and genomics rely upon cultivated clonal cultures, early environmental gene sequencing cloned specific genes (often the 16S rRNA gene) to produce a profile of diversity in a natural sample. Such work revealed that the vast majority of microbial biodiversity had been missed by cultivation-based methods. Because of its ability to reveal the previously hidden diversity of microscopic life, metagenomics offers a powerful lens for viewing the microbial world that has the potential to revolutionize understanding of the entire living world. As the price of DNA sequencing continues to fall, metagenomics now allows microbial ecology to be investigated at a much greater scale ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Human Microbiome Project
The Human Microbiome Project (HMP) was a United States National Institutes of Health (NIH) research initiative to improve understanding of the microbiota involved in human health and disease. Launched in 2007, the first phase (HMP1) focused on identifying and characterizing human microbiota. The second phase, known as the Integrative Human Microbiome Project (iHMP) launched in 2014 with the aim of generating resources to characterize the microbiome and elucidating the roles of microbes in health and disease states. The program received $170 million in funding by the NIH Common Fund from 2007 to 2016. Important components of the HMP were culture-independent methods of microbial community characterization, such as metagenomics (which provides a broad genetic perspective on a single microbial community), as well as extensive whole genome sequencing (which provides a "deep" genetic perspective on certain aspects of a given microbial community, ''i.e.'' of individual bacterial specie ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genomes OnLine Database
The Genomes OnLine Database (GOLD) is a web-based resource for comprehensive information regarding genome and metagenome sequencing projects, and their associated metadata, around the world. Since 2011, the GOLD database has been run by the DOE Joint Genome Institute The GOLD database was created in 1997; the first version of the database contained information for 350 sequencing projects, of which 48 had been completely sequenced with their analyses published. GOLD v.5 was released on 28 May 2014. , the GOLD database contains information for 67,879 genome sequencing projects, of which 7,210 have been completed. In order to facilitate comparative analysis between the information in GOLD and other databases (for example, GenBank and the EMBL), GOLD supports the minimum information standards metadata specifications recommended by the Genomic Standards Consortium, in particular, the MIxS (Minimum Information about any (x) Sequence) specification. GOLD also allows the annotation of ge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genomics
Genomics is an interdisciplinary field of biology focusing on the structure, function, evolution, mapping, and editing of genomes. A genome is an organism's complete set of DNA, including all of its genes as well as its hierarchical, three-dimensional structural configuration. In contrast to genetics, which refers to the study of ''individual'' genes and their roles in inheritance, genomics aims at the collective characterization and quantification of ''all'' of an organism's genes, their interrelations and influence on the organism. Genes may direct the production of proteins with the assistance of enzymes and messenger molecules. In turn, proteins make up body structures such as organs and tissues as well as control chemical reactions and carry signals between cells. Genomics also involves the sequencing and analysis of genomes through uses of high throughput DNA sequencing and bioinformatics to assemble and analyze the function and structure of entire genomes. Advances in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |