directed evolution on:
[Wikipedia]
[Google]
[Amazon]
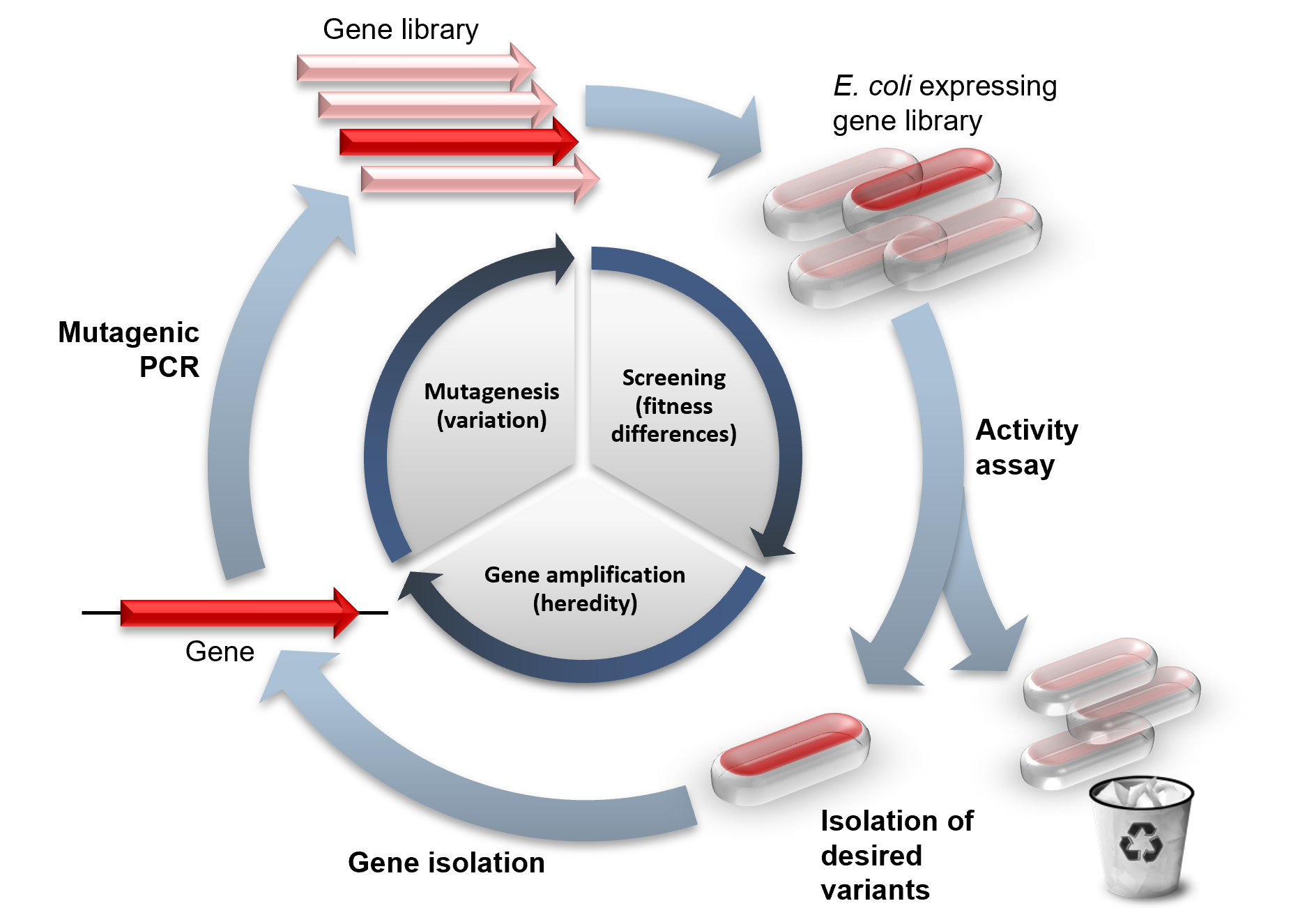 Directed evolution (DE) is a method used in protein engineering that mimics the process of
Directed evolution (DE) is a method used in protein engineering that mimics the process of
 Directed evolution is a mimic of the natural evolution cycle in a laboratory setting. Evolution requires three things to happen: variation between replicators, that the variation causes fitness differences upon which selection acts, and that this variation is
Directed evolution is a mimic of the natural evolution cycle in a laboratory setting. Evolution requires three things to happen: variation between replicators, that the variation causes fitness differences upon which selection acts, and that this variation is
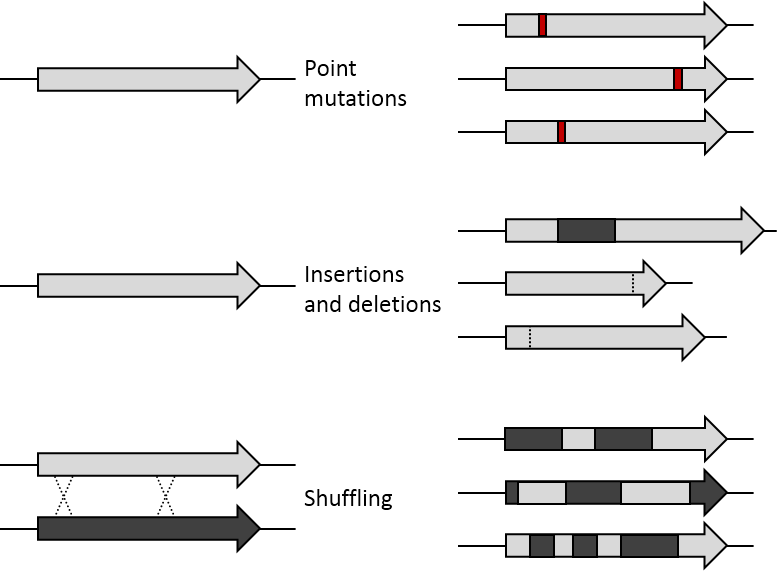 The first step in performing a cycle of directed evolution is the generation of a library of variant genes. The
The first step in performing a cycle of directed evolution is the generation of a library of variant genes. The
 When functional proteins have been isolated, it is necessary that their genes are too, therefore a genotype–phenotype link is required. This can be covalent, such as mRNA display where the
When functional proteins have been isolated, it is necessary that their genes are too, therefore a genotype–phenotype link is required. This can be covalent, such as mRNA display where the
The Dan Tawfik Research Group
*
The Ulrich Schwaneberg Research Group
*
The Frances Arnold Research Group
*
The Huimin Zhao Research Group
*
The Manfred Reetz Research Group
*
*
*
The Chang Liu Research Group
*
The David Liu Research Group
*
The Douglas Clark Research Group
*
*
SeSaM-Biotech – Directed Evolution
Prof. Reetz explains the principle of Directed Evolution
Codexis, Inc.
{{Authority control Evolutionary biology
 Directed evolution (DE) is a method used in protein engineering that mimics the process of
Directed evolution (DE) is a method used in protein engineering that mimics the process of natural selection
Natural selection is the differential survival and reproduction of individuals due to differences in phenotype. It is a key mechanism of evolution, the change in the Heredity, heritable traits characteristic of a population over generation ...
to steer proteins
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, re ...
or nucleic acid
Nucleic acids are large biomolecules that are crucial in all cells and viruses. They are composed of nucleotides, which are the monomer components: a pentose, 5-carbon sugar, a phosphate group and a nitrogenous base. The two main classes of nuclei ...
s toward a user-defined goal. It consists of subjecting a gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei ...
to iterative rounds of mutagenesis
Mutagenesis () is a process by which the genetic information of an organism is changed by the production of a mutation. It may occur spontaneously in nature, or as a result of exposure to mutagens. It can also be achieved experimentally using lab ...
(creating a library of variants), selection (expressing those variants and isolating members with the desired function) and amplification (generating a template for the next round). It can be performed ''in vivo
Studies that are ''in vivo'' (Latin for "within the living"; often not italicized in English) are those in which the effects of various biological entities are tested on whole, living organisms or cells, usually animals, including humans, an ...
'' (in living organisms), or ''in vitro
''In vitro'' (meaning ''in glass'', or ''in the glass'') Research, studies are performed with Cell (biology), cells or biological molecules outside their normal biological context. Colloquially called "test-tube experiments", these studies in ...
'' (in cells or free in solution). Directed evolution is used both for protein engineering as an alternative to rationally designing modified proteins, as well as for experimental evolution studies of fundamental evolutionary principles in a controlled, laboratory environment.
History
Directed evolution has its origins in the 1960s with the evolution of RNA molecules in the "Spiegelman's Monster
Spiegelman's Monster is an RNA chain of only 218 nucleotides that is able to be reproduced by the RNA replication enzyme RNA-dependent RNA polymerase, also called RNA replicase. It is named after its creator, Sol Spiegelman, of the University of I ...
" experiment. The concept was extended to protein evolution via evolution of bacteria under selection pressures that favoured the evolution of a single gene in its genome.
Early phage display
Phage display is a laboratory technique for the study of protein–protein, protein–peptide, and protein–DNA interactions that uses bacteriophages (viruses that infect bacteria) to connect proteins with the genetic information that encodes ...
techniques in the 1980s allowed targeting of mutations and selection to a single protein. This enabled selection of enhanced binding proteins, but was not yet compatible with selection for catalytic activity of enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
s. Methods to evolve enzymes were developed in the 1990s and brought the technique to a wider scientific audience. The field rapidly expanded with new methods for making libraries of gene variants and for screening their activity. The development of directed evolution methods was honored in 2018 with the awarding of the Nobel Prize in Chemistry
The Nobel Prize in Chemistry () is awarded annually by the Royal Swedish Academy of Sciences to scientists in the various fields of chemistry. It is one of the five Nobel Prizes established by the will of Alfred Nobel in 1895, awarded for outst ...
to Frances Arnold
Frances Hamilton Arnold (born July 25, 1956) is an American chemical engineer and Nobel Laureate. She is the Linus Pauling Professor of Chemical Engineering, Bioengineering and Biochemistry at the California Institute of Technology (Caltech). I ...
for evolution of enzymes, and George Smith and Gregory Winter for phage display.
Principles
 Directed evolution is a mimic of the natural evolution cycle in a laboratory setting. Evolution requires three things to happen: variation between replicators, that the variation causes fitness differences upon which selection acts, and that this variation is
Directed evolution is a mimic of the natural evolution cycle in a laboratory setting. Evolution requires three things to happen: variation between replicators, that the variation causes fitness differences upon which selection acts, and that this variation is heritable
Heredity, also called inheritance or biological inheritance, is the passing on of Phenotypic trait, traits from parents to their offspring; either through asexual reproduction or sexual reproduction, the offspring cell (biology), cells or orga ...
. In DE, a single gene is evolved by iterative rounds of mutagenesis, selection or screening, and amplification. Rounds of these steps are typically repeated, using the best variant from one round as the template for the next to achieve stepwise improvements.
The likelihood of success in a directed evolution experiment is directly related to the total library size, as evaluating more mutants increases the chances of finding one with the desired properties.
Generating variation
 The first step in performing a cycle of directed evolution is the generation of a library of variant genes. The
The first step in performing a cycle of directed evolution is the generation of a library of variant genes. The sequence space
In functional analysis and related areas of mathematics, a sequence space is a vector space whose elements are infinite sequences of real or complex numbers. Equivalently, it is a function space whose elements are functions from the natural num ...
for random sequence is vast (10130 possible sequences for a 100 amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
protein) and extremely sparsely populated by functional proteins. Neither experimental, nor natural evolution can ever get close to sampling so many sequences. Of course, natural evolution samples variant sequences close to functional protein sequences and this is imitated in DE by mutagenising an already functional gene.
Some calculations suggest it is entirely feasible that for all practical (i.e. functional and structural) purposes, protein sequence space has been fully explored during the course of evolution of life on Earth.
The starting gene can be mutagenised by random point mutations (by chemical mutagens or error prone PCR) and insertions and deletions (by transposons). Gene recombination can be mimicked by DNA shuffling of several sequences (usually of more than 70% sequence identity) to jump into regions of sequence space between the shuffled parent genes. Finally, specific regions of a gene can be systematically randomised for a more focused approach based on structure and function knowledge. Depending on the method, the library generated will vary in the proportion of functional variants it contains. Even if an organism is used to express the gene of interest, by mutagenising only that gene the rest of the organism's genome remains the same and can be ignored for the evolution experiment (to the extent of providing a constant genetic environment).
Detecting fitness differences
The majority ofmutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, ...
s are deleterious and so libraries of mutants tend to mostly have variants with reduced activity. Therefore, a high-throughput assay
An assay is an investigative (analytic) procedure in laboratory medicine, mining, pharmacology, environmental biology and molecular biology for qualitatively assessing or quantitatively measuring the presence, amount, or functional activity ...
is vital for measuring activity to find the rare variants with beneficial mutations that improve the desired properties. Two main categories of method exist for isolating functional variants. Selection systems directly couple protein function to survival of the gene, whereas screening systems individually assay each variant and allow a quantitative threshold to be set for sorting a variant or population of variants of a desired activity. Both selection and screening can be performed in living cells (''in vivo'' evolution) or performed directly on the protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
or RNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA and deoxyrib ...
without any cells (''in vitro'' evolution).
During ''in vivo'' evolution, each cell (usually bacteria
Bacteria (; : bacterium) are ubiquitous, mostly free-living organisms often consisting of one Cell (biology), biological cell. They constitute a large domain (biology), domain of Prokaryote, prokaryotic microorganisms. Typically a few micr ...
or yeast
Yeasts are eukaryotic, single-celled microorganisms classified as members of the fungus kingdom (biology), kingdom. The first yeast originated hundreds of millions of years ago, and at least 1,500 species are currently recognized. They are est ...
) is transformed with a plasmid
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria and ...
containing a different member of the variant library. In this way, only the gene of interest differs between the cells, with all other genes being kept the same. The cells express the protein either in their cytoplasm
The cytoplasm describes all the material within a eukaryotic or prokaryotic cell, enclosed by the cell membrane, including the organelles and excluding the nucleus in eukaryotic cells. The material inside the nucleus of a eukaryotic cell a ...
or surface
A surface, as the term is most generally used, is the outermost or uppermost layer of a physical object or space. It is the portion or region of the object that can first be perceived by an observer using the senses of sight and touch, and is ...
where its function can be tested. This format has the advantage of selecting for properties in a cellular environment, which is useful when the evolved protein or RNA is to be used in living organisms. When performed without cells, DE involves using ''in vitro'' transcription translation to produce proteins or RNA free in solution or compartmentalised in artificial microdroplets. This method has the benefits of being more versatile in the selection conditions (e.g. temperature, solvent), and can express proteins that would be toxic to cells. Furthermore, ''in vitro'' evolution experiments can generate far larger libraries (up to 1015) because the library DNA need not be inserted into cells (often a limiting step).
Selection
Selection for binding activity is conceptually simple. The target molecule is immobilised on a solid support, a library of variant proteins is flowed over it, poor binders are washed away, and the remaining bound variants recovered to isolate their genes. Binding of an enzyme to immobilised covalentinhibitor
Inhibitor or inhibition may refer to:
Biology
* Enzyme inhibitor, a substance that binds to an enzyme and decreases the enzyme's activity
* Reuptake inhibitor, a substance that increases neurotransmission by blocking the reuptake of a neurotransmi ...
has been also used as an attempt to isolate active catalysts. This approach, however, only selects for single catalytic turnover and is not a good model of substrate binding or true substrate reactivity. If an enzyme activity can be made necessary for cell survival, either by synthesizing a vital metabolite, or destroying a toxin, then cell survival is a function of enzyme activity. Such systems are generally only limited in throughput by the transformation efficiency of cells. They are also less expensive and labour-intensive than screening, however they are typically difficult to engineer, prone to artefacts and give no information on the range of activities present in the library.
Screening
An alternative to selection is a screening system. Each variant gene is individually expressed andassay
An assay is an investigative (analytic) procedure in laboratory medicine, mining, pharmacology, environmental biology and molecular biology for qualitatively assessing or quantitatively measuring the presence, amount, or functional activity ...
ed to quantitatively measure the activity (most often by a colourgenic or fluorogenic product). The variants are then ranked and the experimenter decides which variants to use as templates for the next round of DE. Even the most high throughput assays usually have lower coverage than selection methods but give the advantage of producing detailed information on each one of the screened variants. This disaggregated data can also be used to characterise the distribution of activities in libraries which is not possible in simple selection systems. Screening systems, therefore, have advantages when it comes to experimentally characterising adaptive evolution and fitness landscapes.
Ensuring heredity
 When functional proteins have been isolated, it is necessary that their genes are too, therefore a genotype–phenotype link is required. This can be covalent, such as mRNA display where the
When functional proteins have been isolated, it is necessary that their genes are too, therefore a genotype–phenotype link is required. This can be covalent, such as mRNA display where the mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein.
mRNA is ...
gene is linked to the protein at the end of translation by puromycin. Alternatively the protein and its gene can be co-localised by compartmentalisation in living cells or emulsion droplets. The gene sequences isolated are then amplified by PCR or by transformed host bacteria. Either the single best sequence, or a pool of sequences can be used as the template for the next round of mutagenesis. The repeated cycles of Diversification-Selection-Amplification generate protein variants adapted to the applied selection pressures.
Comparison to rational protein design
Advantages of directed evolution
Rational design
In chemical biology and biomolecular engineering, rational design (RD) is an umbrella term which invites the strategy of creating new molecules with a certain functionality, based upon the ability to predict how the molecule's structure (specific ...
of a protein relies on an in-depth knowledge of the protein structure
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, which are the monomers of the polymer. A single amino acid ...
, as well as its catalytic mechanism
Enzyme catalysis is the increase in the rate of a process by an "enzyme", a biological molecule. Most enzymes are proteins, and most such processes are chemical reactions. Within the enzyme, generally catalysis occurs at a localized site, calle ...
. Specific changes are then made by site-directed mutagenesis in an attempt to change the function of the protein. A drawback of this is that even when the structure and mechanism of action of the protein are well known, the change due to mutation is still difficult to predict. Therefore, an advantage of DE is that there is no need to understand the mechanism of the desired activity or how mutations would affect it.
Limitations of directed evolution
A restriction of directed evolution is that a high-throughput assay is required in order to measure the effects of a large number of different random mutations. This can require extensive research and development before it can be used for directed evolution. Additionally, such assays are often highly specific to monitoring a particular activity and so are not transferable to new DE experiments. Additionally, selecting for improvement in the assayed function simply generates improvements in the assayed function. To understand how these improvements are achieved, the properties of the evolving enzyme have to be measured. Improvement of the assayed activity can be due to improvements in enzyme catalytic activity or enzyme concentration. There is also no guarantee that improvement on one substrate will improve activity on another. This is particularly important when the desired activity cannot be directly screened or selected for and so a ‘proxy’ substrate is used. DE can lead to evolutionary specialisation to the proxy without improving the desired activity. Consequently, choosing appropriate screening or selection conditions is vital for successful DE. The speed of evolution in an experiment also poses a limitation on the utility of directed evolution. For instance, evolution of a particular phenotype, while theoretically feasible, may occur on time-scales that are not practically feasible. Recent theoretical approaches have aimed to overcome the limitation of speed through an application of counter-diabatic driving techniques from statistical physics, though this has yet to be implemented in a directed evolution experiment.Combinatorial approaches
Combined, 'semi-rational' approaches are being investigated to address the limitations of both rational design and directed evolution. Beneficial mutations are rare, so large numbers of random mutants have to be screened to find improved variants. 'Focused libraries' concentrate on randomising regions thought to be richer in beneficial mutations for the mutagenesis step of DE. A focused library contains fewer variants than a traditional random mutagenesis library and so does not require such high-throughput screening. Creating a focused library requires some knowledge of which residues in the structure to mutate. For example, knowledge of theactive site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate, the ''binding s ...
of an enzyme may allow just the residues known to interact with the substrate to be randomised. Alternatively, knowledge of which protein regions are variable in nature can guide mutagenesis in just those regions.
Applications
Directed evolution is frequently used for protein engineering as an alternative torational design
In chemical biology and biomolecular engineering, rational design (RD) is an umbrella term which invites the strategy of creating new molecules with a certain functionality, based upon the ability to predict how the molecule's structure (specific ...
, but can also be used to investigate fundamental questions of enzyme evolution.
Protein engineering
As a protein engineering tool, DE has been most successful in three areas: # Improving protein stability for biotechnological use at high temperatures or in harsh solvents # Improvingbinding affinity
In biochemistry and pharmacology, a ligand is a substance that forms a complex with a biomolecule to serve a biological purpose. The etymology stems from Latin ''ligare'', which means 'to bind'. In protein-ligand binding, the ligand is usuall ...
of therapeutic antibodies (Affinity maturation
In immunology, affinity maturation is the process by which TFH cell-activated B cells produce antibodies with increased affinity for antigen during the course of an immune response. With repeated exposures to the same antigen, a host will produce ...
) and the activity of ''de novo'' designed enzymes
# Altering substrate specificity of existing enzymes, (often for use in industry)
Evolution studies
The study of naturalevolution
Evolution is the change in the heritable Phenotypic trait, characteristics of biological populations over successive generations. It occurs when evolutionary processes such as natural selection and genetic drift act on genetic variation, re ...
is traditionally based on extant organisms and their genes. However, research is fundamentally limited by the lack of fossil
A fossil (from Classical Latin , ) is any preserved remains, impression, or trace of any once-living thing from a past geological age. Examples include bones, shells, exoskeletons, stone imprints of animals or microbes, objects preserve ...
s (and particularly the lack of ancient DNA
Ancient DNA (aDNA) is DNA isolated from ancient sources (typically Biological specimen, specimens, but also environmental DNA). Due to degradation processes (including Crosslinking of DNA, cross-linking, deamination and DNA fragmentation, fragme ...
sequences) and incomplete knowledge of ancient environmental conditions. Directed evolution investigates evolution in a controlled system of genes for individual enzymes
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as pro ...
, ribozymes
Ribozymes (ribonucleic acid enzymes) are RNA molecules that have the ability to catalyze specific biochemical reactions, including RNA splicing in gene expression, similar to the action of protein enzymes. The 1982 discovery of ribozymes demons ...
and replicators (similar to experimental evolution of eukaryotes
The eukaryotes ( ) constitute the domain of Eukaryota or Eukarya, organisms whose cells have a membrane-bound nucleus. All animals, plants, fungi, seaweeds, and many unicellular organisms are eukaryotes. They constitute a major group of ...
, prokaryotes
A prokaryote (; less commonly spelled procaryote) is a single-celled organism whose cell lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Ancient Greek (), meaning 'before', and (), meaning 'nut' ...
and viruses
A virus is a submicroscopic infectious agent that replicates only inside the living cells of an organism. Viruses infect all life forms, from animals and plants to microorganisms, including bacteria and archaea. Viruses are found in almo ...
).
DE allows control of selection pressure, mutation rate
In genetics, the mutation rate is the frequency of new mutations in a single gene, nucleotide sequence, or organism over time. Mutation rates are not constant and are not limited to a single type of mutation; there are many different types of mu ...
and environment (both the abiotic environment such as temperature, and the biotic environment, such as other genes in the organism). Additionally, there is a complete record of all evolutionary intermediate genes. This allows for detailed measurements of evolutionary processes, for example epistasis
Epistasis is a phenomenon in genetics in which the effect of a gene mutation is dependent on the presence or absence of mutations in one or more other genes, respectively termed modifier genes. In other words, the effect of the mutation is depe ...
, evolvability, adaptive constraint fitness landscapes, and neutral networks.
Adaptive laboratory evolution of microbial proteomes
The naturalamino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
composition of proteome
A proteome is the entire set of proteins that is, or can be, expressed by a genome, cell, tissue, or organism at a certain time. It is the set of expressed proteins in a given type of cell or organism, at a given time, under defined conditions. P ...
s can be changed by global canonical amino acids substitutions with suitable noncanonical counterparts under the experimentally imposed selective pressure. For example, global proteome-wide substitutions of natural amino acids with fluorinated analogs have been attempted in ''Escherichia coli'' and ''Bacillus subtilis''. A complete tryptophan
Tryptophan (symbol Trp or W)
is an α-amino acid that is used in the biosynthesis of proteins. Tryptophan contains an α-amino group, an α-carboxylic acid group, and a side chain indole, making it a polar molecule with a non-polar aromat ...
substitution with thienopyrrole-alanine in response to 20899 UGG codon
Genetic code is a set of rules used by living cells to translate information encoded within genetic material (DNA or RNA sequences of nucleotide triplets or codons) into proteins. Translation is accomplished by the ribosome, which links prote ...
s in ''Escherichia coli'' was reported in 2015 by Budisa and Söll. The experimental evolution of microbial strains with a clear-cut accommodation of an additional amino acid is expected to be instrumental for widening the genetic code
Genetic code is a set of rules used by living cell (biology), cells to Translation (biology), translate information encoded within genetic material (DNA or RNA sequences of nucleotide triplets or codons) into proteins. Translation is accomplished ...
experimentally. Directed evolution typically targets a particular gene for mutagenesis
Mutagenesis () is a process by which the genetic information of an organism is changed by the production of a mutation. It may occur spontaneously in nature, or as a result of exposure to mutagens. It can also be achieved experimentally using lab ...
and then screens the resulting variants for a phenotype
In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology (physical form and structure), its developmental processes, its biochemical and physiological propert ...
of interest, often independent of fitness effects, whereas adaptive laboratory evolution selects many genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as ...
-wide mutations that contribute to the fitness of actively growing cultures.
See also
* Applications: ** Protein engineering ** Enzyme engineering ** Protein design ** Expanded genetic code ** Xenobiology * Mutagenesis: ** Random mutagenesis ** Saturated mutagenesis **Staggered extension process The staggered extension process (also referred to as StEP) is a common technique used in biotechnology and molecular biology to create new, mutated genes with qualities of one or more initial genes.
The technique itself is a modified polymerase cha ...
* Selection and screening:
** Yeast display
** Bacterial display
** Phage display
Phage display is a laboratory technique for the study of protein–protein, protein–peptide, and protein–DNA interactions that uses bacteriophages (viruses that infect bacteria) to connect proteins with the genetic information that encodes ...
** Ribosome display
** mRNA display
** FACS
References
External links
* Research groups *The Dan Tawfik Research Group
*
The Ulrich Schwaneberg Research Group
*
The Frances Arnold Research Group
*
The Huimin Zhao Research Group
*
The Manfred Reetz Research Group
*
*
*
The Chang Liu Research Group
*
The David Liu Research Group
*
The Douglas Clark Research Group
*
*
SeSaM-Biotech – Directed Evolution
Prof. Reetz explains the principle of Directed Evolution
Codexis, Inc.
{{Authority control Evolutionary biology