|
Yeast Display
Yeast display (or yeast surface display) is a protein engineering technique that uses the expression of recombinant proteins incorporated into the cell wall of yeast for isolating and engineering antibodies. Development The yeast display technique was first published by the laboratory of Professor K. Dane Wittrup and Eric T. Boder. The technology was sold to Abbott Laboratories in 2001. How it works A protein of interest is displayed as a fusion to the Aga2p protein on the surface of yeast. The Aga2p protein is naturally used by yeast to mediate cell–cell contacts during yeast cell mating. As such, display of a protein via Aga2p projects the protein away from the cell surface, minimizing potential interactions with other molecules on the yeast cell wall. The use of magnetic separation and flow cytometry in conjunction with a yeast display library is a highly effective method to isolate high affinity protein ligands against nearly any receptor through directed evolution. Advant ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Engineering
Protein engineering is the process of developing useful or valuable proteins. It is a young discipline, with much research taking place into the understanding of protein folding and recognition for protein design principles. It has been used to improve the function of many enzymes for industrial catalysis. It is also a product and services market, with an estimated value of $168 billion by 2017. There are two general strategies for protein engineering: rational protein design and directed evolution. These methods are not mutually exclusive; researchers will often apply both. In the future, more detailed knowledge of protein structure and function, and advances in high-throughput screening, may greatly expand the abilities of protein engineering. Eventually, even unnatural amino acids may be included, via newer methods, such as expanded genetic code, that allow encoding novel amino acids in genetic code. Approaches Rational design In rational protein design, a scientist uses ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
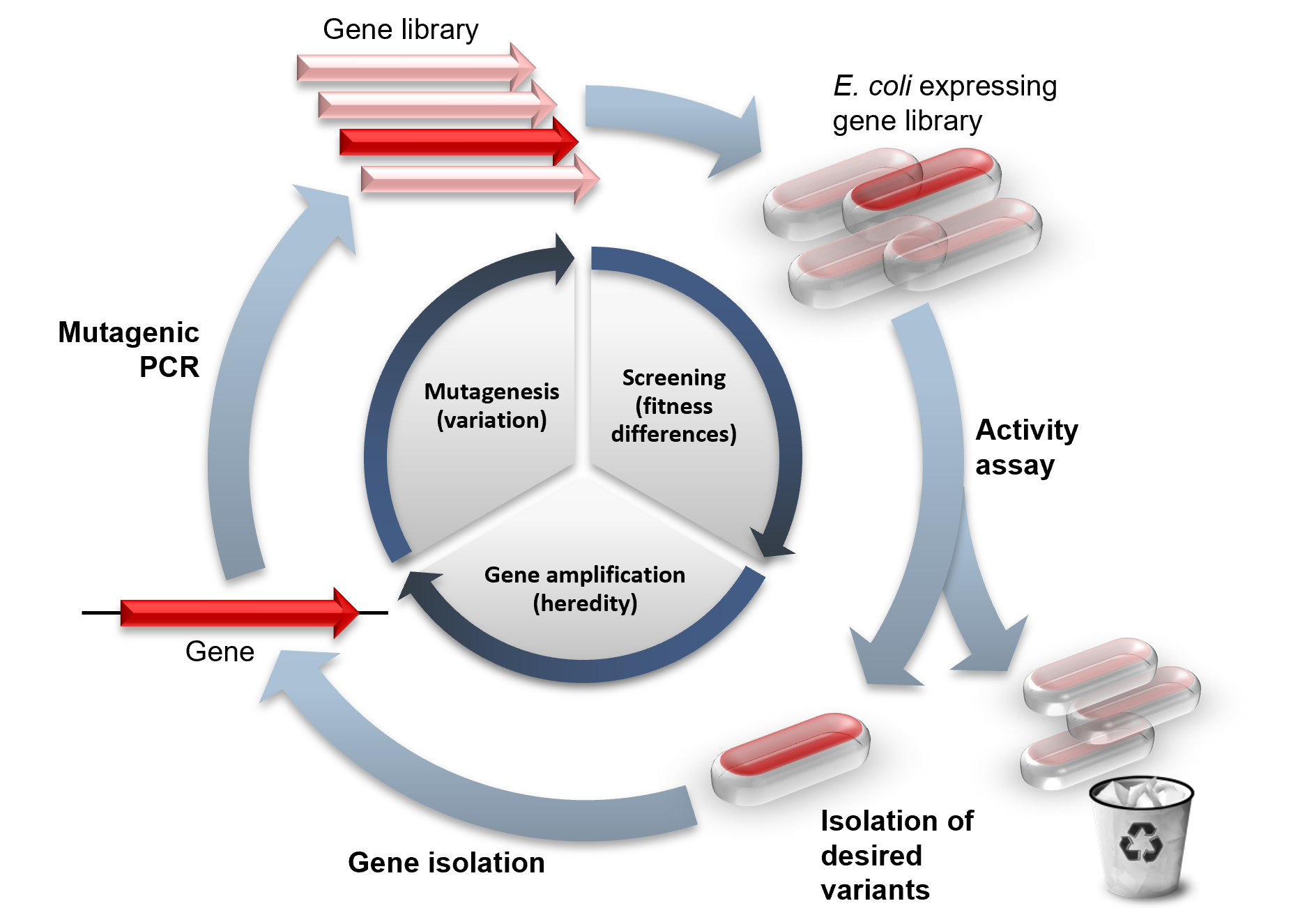
Directed Evolution
Directed evolution (DE) is a method used in protein engineering that mimics the process of natural selection to steer proteins or nucleic acids toward a user-defined goal. It consists of subjecting a gene to iterative rounds of mutagenesis (creating a library of variants), selection (expressing those variants and isolating members with the desired function) and amplification (generating a template for the next round). It can be performed ''in vivo'' (in living organisms), or ''in vitro'' (in cells or free in solution). Directed evolution is used both for protein engineering as an alternative to rationally designing modified proteins, as well as for experimental evolution studies of fundamental evolutionary principles in a controlled, laboratory environment. History Directed evolution has its origins in the 1960s with the evolution of RNA molecules in the " Spiegelman's Monster" experiment. The concept was extended to protein evolution via evolution of bacteria under select ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacterial Display
Bacterial display (or bacteria display or bacterial surface display) is a protein engineering technique used for in vitro protein evolution. Libraries of polypeptides displayed on the surface of bacteria can be screened using flow cytometry or iterative selection procedures (biopanning). This protein engineering technique allows us to link the function of a protein with the gene that encodes it. Bacterial display can be used to find target proteins with desired properties and can be used to make affinity ligands which are cell-specific. This system can be used in many applications including the creation of novel vaccines, the identification of enzyme substrates and finding the affinity of a ligand for its target protein. Bacterial display is often coupled with magnetic-activated cell sorting (MACS) or fluorescence-activated cell sorting (FACS) techniques. Competing methods for protein evolution ''in vitro'' are phage display, ribosome display, yeast display, and mRNA display. Bacter ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosome Display
Ribosome display is a technique used to perform ''in vitro'' protein evolution to create proteins that can bind to a desired ligand. The process results in translated proteins that are associated with their mRNA progenitor which is used, as a complex, to bind to an immobilized ligand in a selection step. The mRNA-protein hybrids that bind well are then reverse transcribed to cDNA and their sequence amplified via PCR. The result is a nucleotide sequence that can be used to create tightly binding proteins. Ribosome display process Ribosome display begins with a native library of DNA sequences coding for polypeptides. Each sequence is transcribed, and then translated ''in vitro'' into polypeptide. However, the DNA library coding for a particular library of binding proteins is genetically fused to a spacer sequence lacking a stop codon before its end. The lack of a stop codon prevents release factors from binding and triggering the disassembly of the translational complex. So, thi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phage Display
Phage display is a laboratory technique for the study of protein–protein, protein–peptide, and protein– DNA interactions that uses bacteriophages (viruses that infect bacteria) to connect proteins with the genetic information that encodes them. In this technique, a gene encoding a protein of interest is inserted into a phage coat protein gene, causing the phage to "display" the protein on its outside while containing the gene for the protein on its inside, resulting in a connection between genotype and phenotype. These displaying phages can then be screened against other proteins, peptides or DNA sequences, in order to detect interaction between the displayed protein and those other molecules. In this way, large libraries of proteins can be screened and amplified in a process called ''in vitro'' selection, which is analogous to natural selection. The most common bacteriophages used in phage display are M13 and fd filamentous phage, though T4, T7, and λ phage have ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mammalian Display
Mammals () are a group of vertebrate animals constituting the class Mammalia (), characterized by the presence of mammary glands which in females produce milk for feeding (nursing) their young, a neocortex (a region of the brain), fur or hair, and three middle ear bones. These characteristics distinguish them from reptiles (including birds) from which they diverged in the Carboniferous, over 300 million years ago. Around 6,400 extant species of mammals have been described divided into 29 orders. The largest orders, in terms of number of species, are the rodents, bats, and Eulipotyphla (hedgehogs, moles, shrews, and others). The next three are the Primates (including humans, apes, monkeys, and others), the Artiodactyla (cetaceans and even-toed ungulates), and the Carnivora (cats, dogs, seals, and others). In terms of cladistics, which reflects evolutionary history, mammals are the only living members of the Synapsida (synapsids); this clade, together with Sauropsida ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glycosylation
Glycosylation is the reaction in which a carbohydrate (or ' glycan'), i.e. a glycosyl donor, is attached to a hydroxyl or other functional group of another molecule (a glycosyl acceptor) in order to form a glycoconjugate. In biology (but not always in chemistry), glycosylation usually refers to an enzyme-catalysed reaction, whereas glycation (also 'non-enzymatic glycation' and 'non-enzymatic glycosylation') may refer to a non-enzymatic reaction (though in practice, 'glycation' often refers more specifically to Maillard-type reactions). Glycosylation is a form of co-translational and post-translational modification. Glycans serve a variety of structural and functional roles in membrane and secreted proteins. The majority of proteins synthesized in the rough endoplasmic reticulum undergo glycosylation. Glycosylation is also present in the cytoplasm and nucleus as the ''O''-GlcNAc modification. Aglycosylation is a feature of engineered antibodies to bypass glycosylation. Five clas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fluorescent-activated Cell Sorting
Flow cytometry (FC) is a technique used to detect and measure physical and chemical characteristics of a population of cells or particles. In this process, a sample containing cells or particles is suspended in a fluid and injected into the flow cytometer instrument. The sample is focused to ideally flow one cell at a time through a laser beam, where the light scattered is characteristic to the cells and their components. Cells are often labeled with fluorescent markers so light is absorbed and then emitted in a band of wavelengths. Tens of thousands of cells can be quickly examined and the data gathered are processed by a computer. Flow cytometry is routinely used in basic research, clinical practice, and clinical trials. Uses for flow cytometry include: * Cell counting * Cell sorting * Determining cell characteristics and function * Detecting microorganisms * Biomarker detection * Protein engineering detection * Diagnosis of health disorders such as blood cancers * Measuring ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Receptor (biochemistry)
In biochemistry and pharmacology, receptors are chemical structures, composed of protein, that receive and transduce signals that may be integrated into biological systems. These signals are typically chemical messengers which bind to a receptor and cause some form of cellular/tissue response, e.g. a change in the electrical activity of a cell. There are three main ways the action of the receptor can be classified: relay of signal, amplification, or integration. Relaying sends the signal onward, amplification increases the effect of a single ligand, and integration allows the signal to be incorporated into another biochemical pathway. Receptor proteins can be classified by their location. Transmembrane receptors include ligand-gated ion channels, G protein-coupled receptors, and enzyme-linked hormone receptors. Intracellular receptors are those found inside the cell, and include cytoplasmic receptors and nuclear receptors. A molecule that binds to a receptor is called a ligand ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Recombinant Protein
Recombinant DNA (rDNA) molecules are DNA molecules formed by laboratory methods of genetic recombination (such as molecular cloning) that bring together genetic material from multiple sources, creating sequences that would not otherwise be found in the genome. Recombinant DNA is the general name for a piece of DNA that has been created by combining at least two fragments from two different sources. Recombinant DNA is possible because DNA molecules from all organisms share the same chemical structure, and differ only in the nucleotide sequence within that identical overall structure. Recombinant DNA molecules are sometimes called chimeric DNA, because they can be made of material from two different species, like the mythical chimera. R-DNA technology uses palindromic sequences and leads to the production of sticky and blunt ends. The DNA sequences used in the construction of recombinant DNA molecules can originate from any species. For example, plant DNA may be joined to bacte ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ligand (biochemistry)
In biochemistry and pharmacology, a ligand is a substance that forms a complex with a biomolecule to serve a biological purpose. The etymology stems from ''ligare'', which means 'to bind'. In protein-ligand binding, the ligand is usually a molecule which produces a signal by binding to a site on a target protein. The binding typically results in a change of conformational isomerism (conformation) of the target protein. In DNA-ligand binding studies, the ligand can be a small molecule, ion, or protein which binds to the DNA double helix. The relationship between ligand and binding partner is a function of charge, hydrophobicity, and molecular structure. Binding occurs by intermolecular forces, such as ionic bonds, hydrogen bonds and Van der Waals forces. The association or docking is actually reversible through dissociation. Measurably irreversible covalent bonding between a ligand and target molecule is atypical in biological systems. In contrast to the definition of lig ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |