|
Saturated Mutagenesis
Site saturation mutagenesis (SSM), or simply site saturation, is a random mutagenesis technique used in protein engineering, in which a single codon The genetic code is the set of rules used by living cells to translate information encoded within genetic material ( DNA or RNA sequences of nucleotide triplets, or codons) into proteins. Translation is accomplished by the ribosome, which links ... or set of codons is substituted with all possible amino acids at the position. There are many variants of the site saturation technique, from paired site saturation (saturating two positions in every mutant in the library) to scanning site saturation (performing a site saturation at every site in the protein, resulting in a library of size 0 x (number of residues in the protein)that contains every possible point mutant of the protein). Method Saturation mutagenesis is commonly achieved by site-directed mutagenesis PCR with a randomised codon in the primers (e.g. SeSaM) or by a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Site Saturation Mutagenesis
Site saturation mutagenesis (SSM), or simply site saturation, is a Mutagenesis (molecular biology technique), random mutagenesis technique used in protein engineering, in which a single codon or set of codons is substituted with all possible amino acids at the position. There are many variants of the site saturation technique, from paired site saturation (saturating two positions in every mutant in the library) to scanning site saturation (performing a site saturation at every site in the protein, resulting in a library of size [20 x (number of residues in the protein)] that contains every possible point mutation, point mutant of the protein). Method Saturation mutagenesis is commonly achieved by site-directed mutagenesis PCR with a randomised codon in the primers (e.g. SeSaM) or by Artificial gene synthesis#Applications, artificial gene synthesis, with a mixture of synthesis nucleotides used at the codons to be randomised. Different degenerate codons can be used to encode set ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mutagenesis (molecular Biology Technique)
In molecular biology, mutagenesis is an important laboratory technique whereby DNA mutations are deliberately engineered to produce libraries of mutant genes, proteins, strains of bacteria, or other genetically modified organisms. The various constituents of a gene, as well as its regulatory elements and its gene products, may be mutated so that the functioning of a genetic locus, process, or product can be examined in detail. The mutation may produce mutant proteins with interesting properties or enhanced or novel functions that may be of commercial use. Mutant strains may also be produced that have practical application or allow the molecular basis of a particular cell function to be investigated. Many methods of mutagenesis exist today. Initially, the kind of mutations artificially induced in the laboratory were entirely random using mechanisms such as UV irradiation. Random mutagenesis cannot target specific regions or sequences of the genome; however, with the development o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Engineering
Protein engineering is the process of developing useful or valuable proteins. It is a young discipline, with much research taking place into the understanding of protein folding and recognition for protein design principles. It has been used to improve the function of many enzymes for industrial catalysis. It is also a product and services market, with an estimated value of $168 billion by 2017. There are two general strategies for protein engineering: rational protein design and directed evolution. These methods are not mutually exclusive; researchers will often apply both. In the future, more detailed knowledge of protein structure and function, and advances in high-throughput screening, may greatly expand the abilities of protein engineering. Eventually, even unnatural amino acids may be included, via newer methods, such as expanded genetic code, that allow encoding novel amino acids in genetic code. Approaches Rational design In rational protein design, a scientist ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
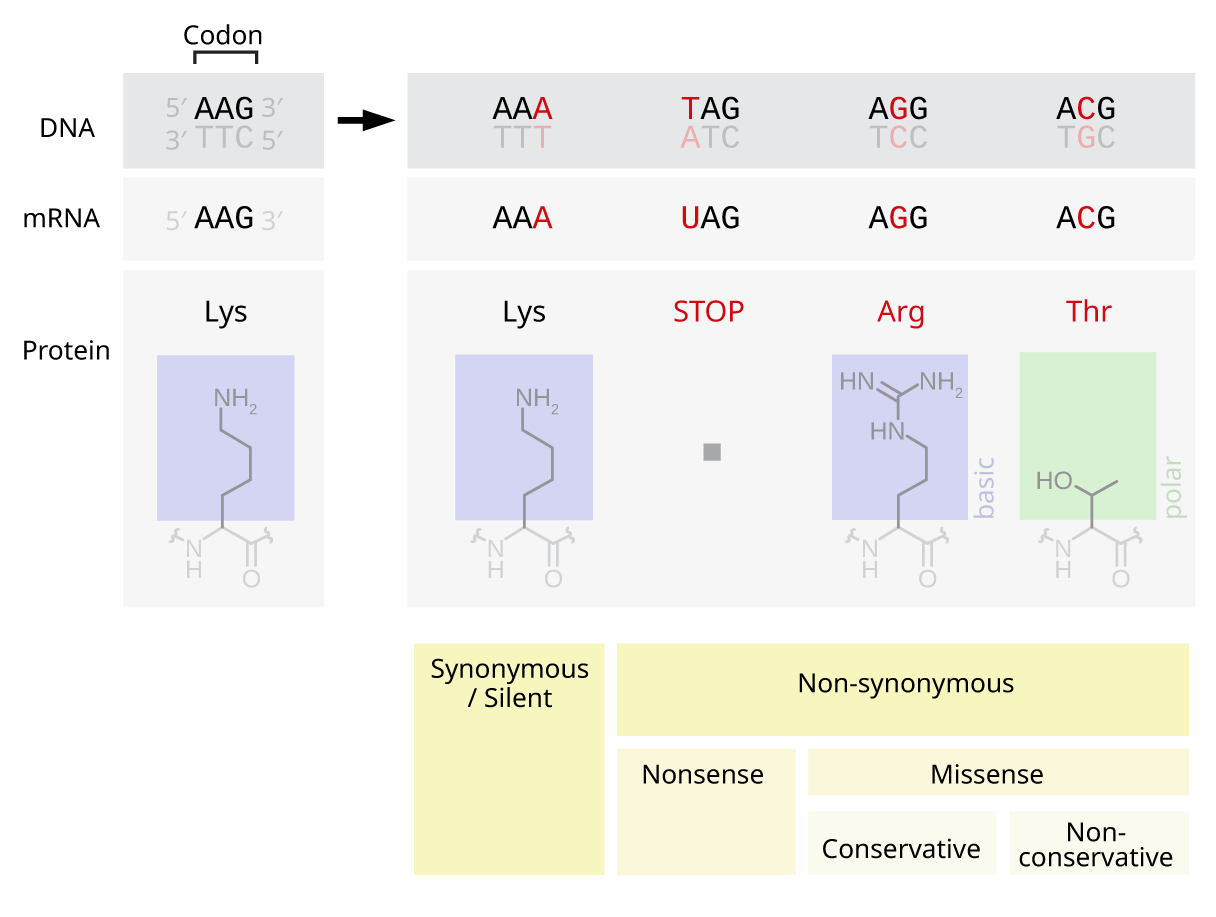
Codon
The genetic code is the set of rules used by living cells to translate information encoded within genetic material ( DNA or RNA sequences of nucleotide triplets, or codons) into proteins. Translation is accomplished by the ribosome, which links proteinogenic amino acids in an order specified by messenger RNA (mRNA), using transfer RNA (tRNA) molecules to carry amino acids and to read the mRNA three nucleotides at a time. The genetic code is highly similar among all organisms and can be expressed in a simple table with 64 entries. The codons specify which amino acid will be added next during protein biosynthesis. With some exceptions, a three-nucleotide codon in a nucleic acid sequence specifies a single amino acid. The vast majority of genes are encoded with a single scheme (see the RNA codon table). That scheme is often referred to as the canonical or standard genetic code, or simply ''the'' genetic code, though variant codes (such as in mitochondria) exist. History Ef ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha amino acids appear in the genetic code. Amino acids can be classified according to the locations of the core structural functional groups, as Alpha and beta carbon, alpha- , beta- , gamma- or delta- amino acids; other categories relate to Chemical polarity, polarity, ionization, and side chain group type (aliphatic, Open-chain compound, acyclic, aromatic, containing hydroxyl or sulfur, etc.). In the form of proteins, amino acid ''residues'' form the second-largest component ( water being the largest) of human muscles and other tissues. Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesis. It is thought that they played a key role in enabling li ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
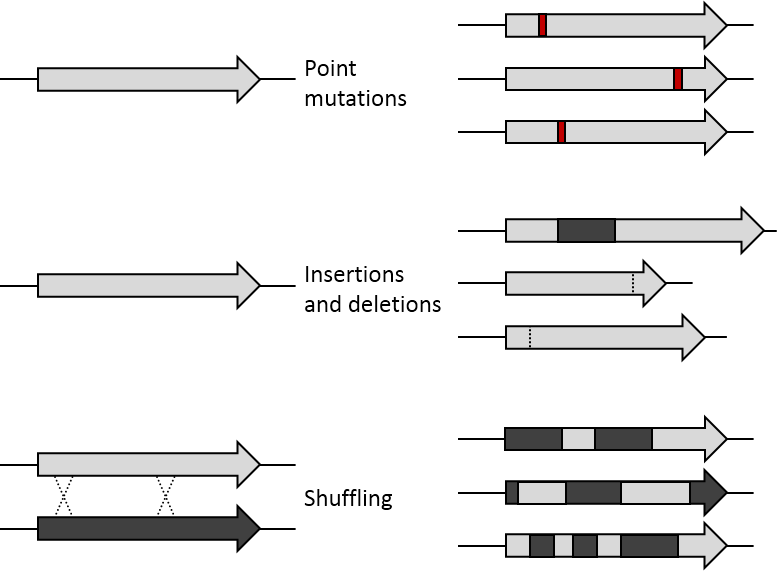
Point Mutation
A point mutation is a genetic mutation where a single nucleotide base is changed, inserted or deleted from a DNA or RNA sequence of an organism's genome. Point mutations have a variety of effects on the downstream protein product—consequences that are moderately predictable based upon the specifics of the mutation. These consequences can range from no effect (e.g. synonymous mutations) to deleterious effects (e.g. frameshift mutations), with regard to protein production, composition, and function. Causes Point mutations usually take place during DNA replication. DNA replication occurs when one double-stranded DNA molecule creates two single strands of DNA, each of which is a template for the creation of the complementary strand. A single point mutation can change the whole DNA sequence. Changing one purine or pyrimidine may change the amino acid that the nucleotides code for. Point mutations may arise from spontaneous mutations that occur during DNA replication. The rate of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Site-directed Mutagenesis
Site-directed mutagenesis is a molecular biology method that is used to make specific and intentional mutating changes to the DNA sequence of a gene and any gene products. Also called site-specific mutagenesis or oligonucleotide-directed mutagenesis, it is used for investigating the structure and biological activity of DNA, RNA, and protein molecules, and for protein engineering. Site-directed mutagenesis is one of the most important laboratory techniques for creating DNA libraries by introducing mutations into DNA sequences. There are numerous methods for achieving site-directed mutagenesis, but with decreasing costs of oligonucleotide synthesis, artificial gene synthesis is now occasionally used as an alternative to site-directed mutagenesis. Since 2013, the development of the CRISPR/Cas9 technology, based on a prokaryotic viral defense system, has also allowed for the editing of the genome, and mutagenesis may be performed ''in vivo'' with relative ease. History Early attempt ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SeSaM ''
{{disambiguation ...
Sesam, SESAM or SeSaM may refer to: * SESAM (database), a relational database developed by Fujitsu Siemens * SESAM (FEM), a structural analysis software * Sesam (search engine), a Scandinavian internet search engine * SeSaM-Biotech GmbH, a biotechnology company focusing on protein engineering * Semiconductor saturable-absorber mirror, a component used in some lasers * Sequence saturation mutagenesis (SeSaM), a molecular biology method for diversity creation by random mutagenesis See also * Sesame (other) * Various wild and cultivated plants in the genus ''Sesamum ''Sesamum'' is a leguminous crop and genus of about 20 species in the flowering plant family Pedaliaceae. The plants are annual or perennial herbs with edible seeds. The best-known member of the genus is sesame, '' Sesamum indicum'' (syn. ''Sesa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Artificial Gene Synthesis
Artificial gene synthesis, or simply gene synthesis, refers to a group of methods that are used in synthetic biology to construct and assemble genes from nucleotides '' de novo''. Unlike DNA synthesis in living cells, artificial gene synthesis does not require template DNA, allowing virtually any DNA sequence to be synthesized in the laboratory. It comprises two main steps, the first of which is solid-phase DNA synthesis, sometimes known as DNA printing. This produces oligonucleotide fragments that are generally under 200 base pairs. The second step then involves connecting these oligonucleotide fragments using various DNA assembly methods. Because artificial gene synthesis does not require template DNA, it is theoretically possible to make a completely synthetic DNA molecule with no limits on the nucleotide sequence or size. Synthesis of the first complete gene, a yeast tRNA, was demonstrated by Har Gobind Khorana and coworkers in 1972. Synthesis of the first peptide- and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleotide
Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver. Nucleotides are composed of three subunit molecules: a nucleobase, a five-carbon sugar ( ribose or deoxyribose), and a phosphate group consisting of one to three phosphates. The four nucleobases in DNA are guanine, adenine, cytosine and thymine; in RNA, uracil is used in place of thymine. Nucleotides also play a central role in metabolism at a fundamental, cellular level. They provide chemical energy—in the form of the nucleoside triphosphates, adenosine triphosphate (ATP), guanosine triphosphate (GTP), cytidine triphosphate (CTP) and uridine triphosphate (UTP)—throughout the cell for the many c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleic Acid Notation
The nucleic acid notation currently in use was first formalized by the International Union of Pure and Applied Chemistry (IUPAC) in 1970. This universally accepted notation uses the Roman characters G, C, A, and T, to represent the four nucleotides commonly found in deoxyribonucleic acids (DNA). Given the rapidly expanding role for genetic sequencing, synthesis, and analysis in biology, some researchers have developed alternate notations to further support the analysis and manipulation of genetic data. These notations generally exploit size, shape, and symmetry to accomplish these objectives. IUPAC notation Degenerate base symbols in biochemistry are an IUPAC representation for a position on a DNA sequence that can have multiple possible alternatives. These should not be confused with non-canonical bases because each particular sequence will have in fact one of the regular bases. These are used to encode the consensus sequence of a population of aligned sequences and are used f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |