|
Polypyrimidine Tract
{{Short description, A pyrimidine-rich sequence involved in pre-messenger RNA maturation The polypyrimidine tract is a region of pre-messenger RNA (mRNA) that promotes the assembly of the spliceosome, the protein complex specialized for carrying out RNA splicing during the process of post-transcriptional modification. The region is rich with pyrimidine nucleotides, especially uracil, and is usually 15–20 base pairs long, located about 5–40 base pairs before the 3' end of the intron to be spliced.Lodish H, Berk A, Matsudaira P, Kaiser CA, Krieger M, Scott MP, Zipursky SL, Darnell J. (2004). ''Molecular Cell Biology.'' WH Freeman: New York, NY. 5th ed. A number of protein factors bind to or associate with the polypyrimidine tract, including the spliceosome component U2AF and the ''polypyrimidine tract-binding protein'' (PTB), which plays a regulatory role in alternative splicing. PTB's primary function is in exon silencing, by which a particular exon region normally spliced into ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
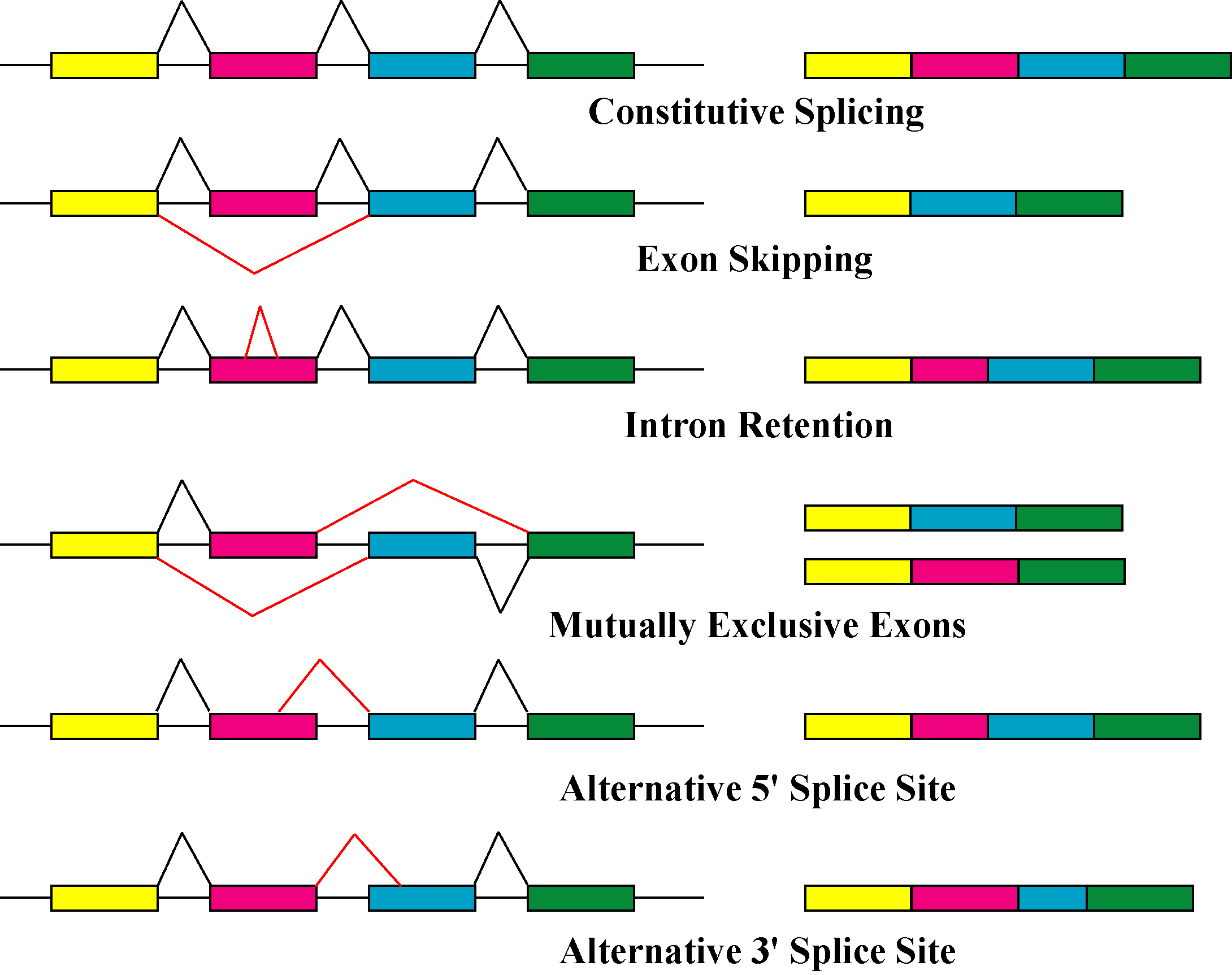
Alternative Splicing
Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during gene expression that allows a single gene to code for multiple proteins. In this process, particular exons of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. This means the exons are joined in different combinations, leading to different (alternative) mRNA strands. Consequently, the proteins translated from alternatively spliced mRNAs will contain differences in their amino acid sequence and, often, in their biological functions (see Figure). Biologically relevant alternative splicing occurs as a normal phenomenon in eukaryotes, where it increases the number of proteins that can be encoded by the genome. In humans, it is widely believed that ~95% of multi-exonic genes are alternatively spliced to produce functional alternative products from the same gene but many scientists believe that most o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
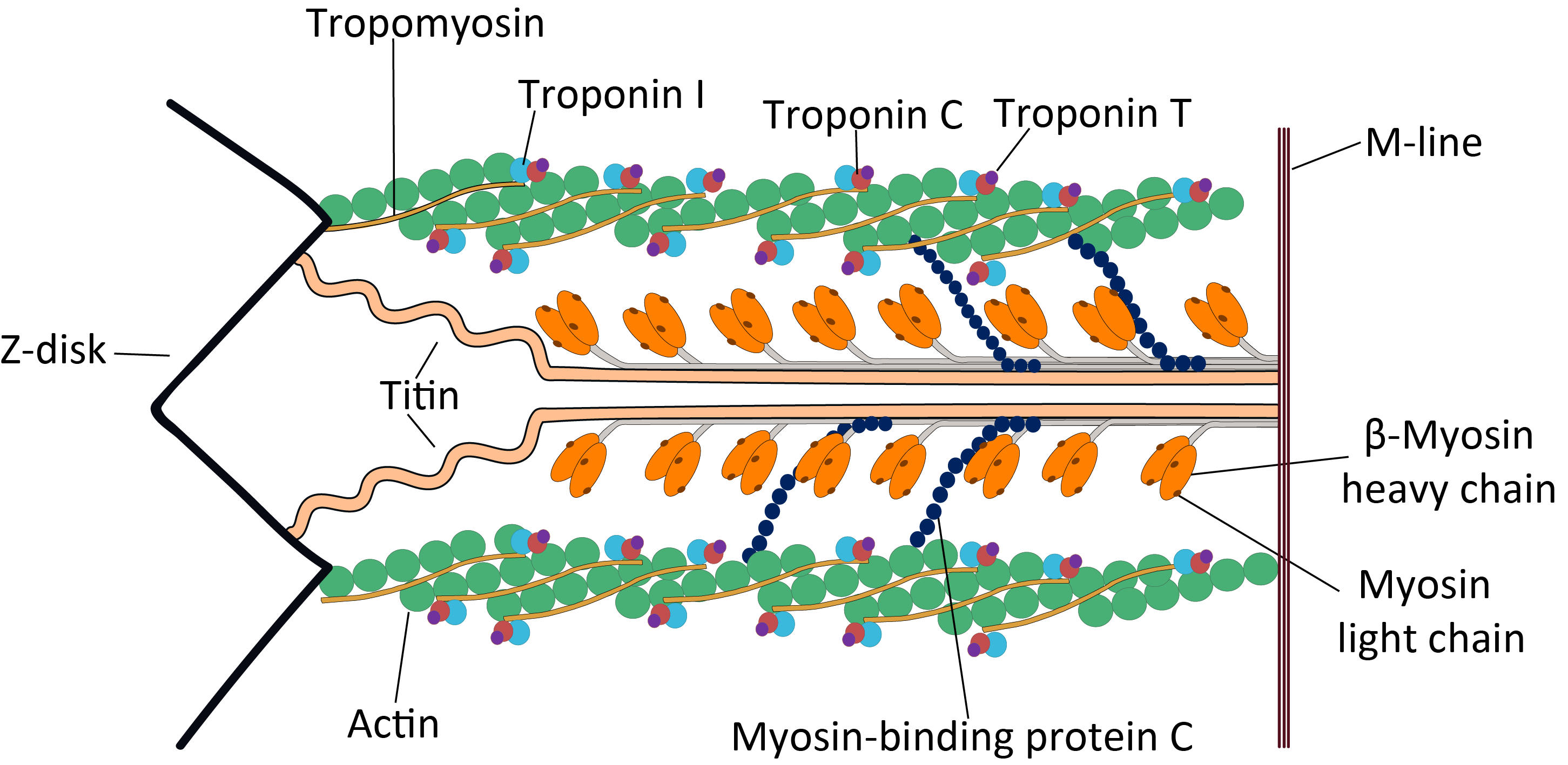
Tropomyosin
Tropomyosin is a two-stranded alpha-helical, coiled coil protein found in actin-based cytoskeletons. Tropomyosin and the actin skeleton All organisms contain organelles that provide physical integrity to their cells. These type of organelles are collectively known as the cytoskeleton, and one of the most ancient systems is based on filamentous polymers of the protein actin. A polymer of a second protein, tropomyosin, is an integral part of most actin filaments in animals. Tropomyosins are a large family of integral components of actin filaments that play a critical role in regulating the function of actin filaments in both muscle and nonmuscle cells. These proteins consist of rod-shaped coiled-coil hetero- or homo- dimers that lie along the α-helical groove of most actin filaments. Interaction occurs along the length of the actin filament, with dimers aligning in a head-to-tail fashion. Tropomyosins are often categorised into two groups, muscle tropomyosin isoforms and nonmus ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
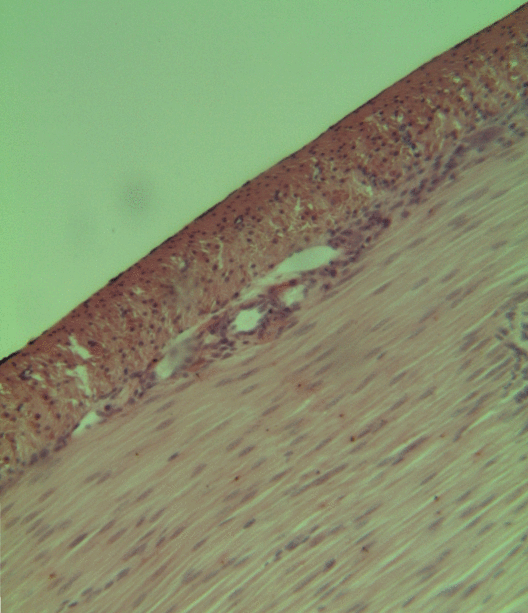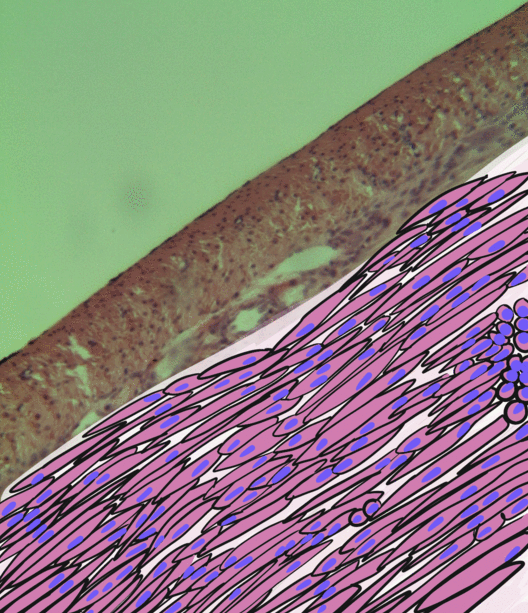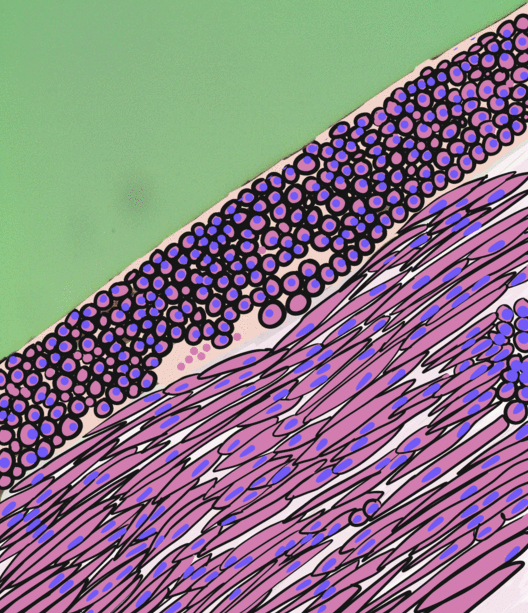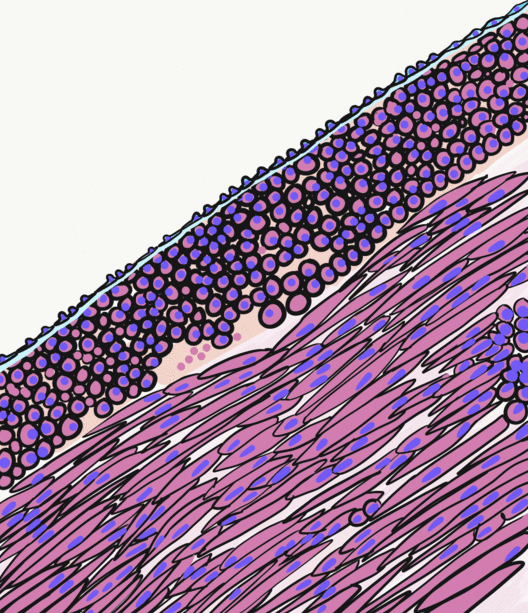
Skeletal Muscle
Skeletal muscles (commonly referred to as muscles) are organs of the vertebrate muscular system and typically are attached by tendons to bones of a skeleton. The muscle cells of skeletal muscles are much longer than in the other types of muscle tissue, and are often known as muscle fibers. The muscle tissue of a skeletal muscle is striated – having a striped appearance due to the arrangement of the sarcomeres. Skeletal muscles are voluntary muscles under the control of the somatic nervous system. The other types of muscle are cardiac muscle which is also striated and smooth muscle which is non-striated; both of these types of muscle tissue are classified as involuntary, or, under the control of the autonomic nervous system. A skeletal muscle contains multiple muscle fascicle, fascicles – bundles of muscle fibers. Each individual fiber, and each muscle is surrounded by a type of connective tissue layer of fascia. Muscle fibers are formed from the cell fusion, fusion of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Smooth Muscle
Smooth muscle is an involuntary non-striated muscle, so-called because it has no sarcomeres and therefore no striations (''bands'' or ''stripes''). It is divided into two subgroups, single-unit and multiunit smooth muscle. Within single-unit muscle, the whole bundle or sheet of smooth muscle cells contracts as a syncytium. Smooth muscle is found in the walls of hollow organs, including the stomach, intestines, bladder and uterus; in the walls of passageways, such as blood, and lymph vessels, and in the tracts of the respiratory, urinary, and reproductive systems. In the eyes, the ciliary muscles, a type of smooth muscle, dilate and contract the iris and alter the shape of the lens. In the skin, smooth muscle cells such as those of the arrector pili cause hair to stand erect in response to cold temperature or fear. Structure Gross anatomy Smooth muscle is grouped into two types: single-unit smooth muscle, also known as visceral smooth muscle, and multiunit smooth muscle. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biological Tissue
In biology, tissue is a biological organizational level between cells and a complete organ. A tissue is an ensemble of similar cells and their extracellular matrix from the same origin that together carry out a specific function. Organs are then formed by the functional grouping together of multiple tissues. The English word "tissue" derives from the French word "tissu", the past participle of the verb tisser, "to weave". The study of tissues is known as histology or, in connection with disease, as histopathology. Xavier Bichat is considered as the "Father of Histology". Plant histology is studied in both plant anatomy and physiology. The classical tools for studying tissues are the paraffin block in which tissue is embedded and then sectioned, the histological stain, and the optical microscope. Developments in electron microscopy, immunofluorescence, and the use of frozen tissue-sections have enhanced the detail that can be observed in tissues. With these tools, the c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Splice Site
RNA splicing is a process in molecular biology where a newly-made precursor messenger RNA (pre-mRNA) transcript is transformed into a mature messenger RNA (mRNA). It works by removing all the introns (non-coding regions of RNA) and ''splicing'' back together exons (coding regions). For nuclear-encoded genes, splicing occurs in the nucleus either during or immediately after transcription. For those eukaryotic genes that contain introns, splicing is usually needed to create an mRNA molecule that can be translated into protein. For many eukaryotic introns, splicing occurs in a series of reactions which are catalyzed by the spliceosome, a complex of small nuclear ribonucleoproteins ( snRNPs). There exist self-splicing introns, that is, ribozymes that can catalyze their own excision from their parent RNA molecule. The process of transcription, splicing and translation is called gene expression, the central dogma of molecular biology. Splicing pathways Several methods of RNA spli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryote
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacteria and Archaea (both prokaryotes) make up the other two domains. The eukaryotes are usually now regarded as having emerged in the Archaea or as a sister of the Asgard archaea. This implies that there are only two domains of life, Bacteria and Archaea, with eukaryotes incorporated among archaea. Eukaryotes represent a small minority of the number of organisms, but, due to their generally much larger size, their collective global biomass is estimated to be about equal to that of prokaryotes. Eukaryotes emerged approximately 2.3–1.8 billion years ago, during the Proterozoic eon, likely as flagellated phagotrophs. Their name comes from the Greek εὖ (''eu'', "well" or "good") and κάρυον (''karyon'', "nut" or "kernel"). Euka ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Code
The genetic code is the set of rules used by living cells to translate information encoded within genetic material ( DNA or RNA sequences of nucleotide triplets, or codons) into proteins. Translation is accomplished by the ribosome, which links proteinogenic amino acids in an order specified by messenger RNA (mRNA), using transfer RNA (tRNA) molecules to carry amino acids and to read the mRNA three nucleotides at a time. The genetic code is highly similar among all organisms and can be expressed in a simple table with 64 entries. The codons specify which amino acid will be added next during protein biosynthesis. With some exceptions, a three-nucleotide codon in a nucleic acid sequence specifies a single amino acid. The vast majority of genes are encoded with a single scheme (see the RNA codon table). That scheme is often referred to as the canonical or standard genetic code, or simply ''the'' genetic code, though variant codes (such as in mitochondria) exist. History Effor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Isoform
A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene or gene family and are the result of genetic differences. While many perform the same or similar biological roles, some isoforms have unique functions. A set of protein isoforms may be formed from alternative splicings, variable promoter usage, or other post-transcriptional modifications of a single gene; post-translational modifications are generally not considered. (For that, see Proteoforms.) Through RNA splicing mechanisms, mRNA has the ability to select different protein-coding segments ( exons) of a gene, or even different parts of exons from RNA to form different mRNA sequences. Each unique sequence produces a specific form of a protein. The discovery of isoforms could explain the discrepancy between the small number of protein coding regions genes revealed by the human genome project and the large diversity of proteins seen in an organism: different ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication. The process of gene expression is used by all known life—eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea), and utilized by viruses—to generate the macromolecular machinery for life. In genetics, gene expression is the most fundamental level at which the genotype gives rise to the phenotype, '' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Exon
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequence in RNA transcripts. In RNA splicing, introns are removed and exons are covalently joined to one another as part of generating the mature RNA. Just as the entire set of genes for a species constitutes the genome, the entire set of exons constitutes the exome. History The term ''exon'' derives from the expressed region and was coined by American biochemist Walter Gilbert in 1978: "The notion of the cistron… must be replaced by that of a transcription unit containing regions which will be lost from the mature messengerwhich I suggest we call introns (for intragenic regions)alternating with regions which will be expressedexons." This definition was originally made for protein-coding transcripts that are spliced before being translated. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |