Eukaryote on:
[Wikipedia]
[Google]
[Amazon]
Eukaryotes () are
 Eukaryote cells include a variety of membrane-bound structures, collectively referred to as the endomembrane system. Simple compartments, called
Eukaryote cells include a variety of membrane-bound structures, collectively referred to as the endomembrane system. Simple compartments, called

 Many eukaryotes have long slender motile cytoplasmic projections, called flagella, or similar structures called cilia. Flagella and cilia are sometimes referred to as
Many eukaryotes have long slender motile cytoplasmic projections, called flagella, or similar structures called cilia. Flagella and cilia are sometimes referred to as

 All animals are eukaryotic. Animal cells are distinct from those of other eukaryotes, most notably plants, as they lack
All animals are eukaryotic. Animal cells are distinct from those of other eukaryotes, most notably plants, as they lack
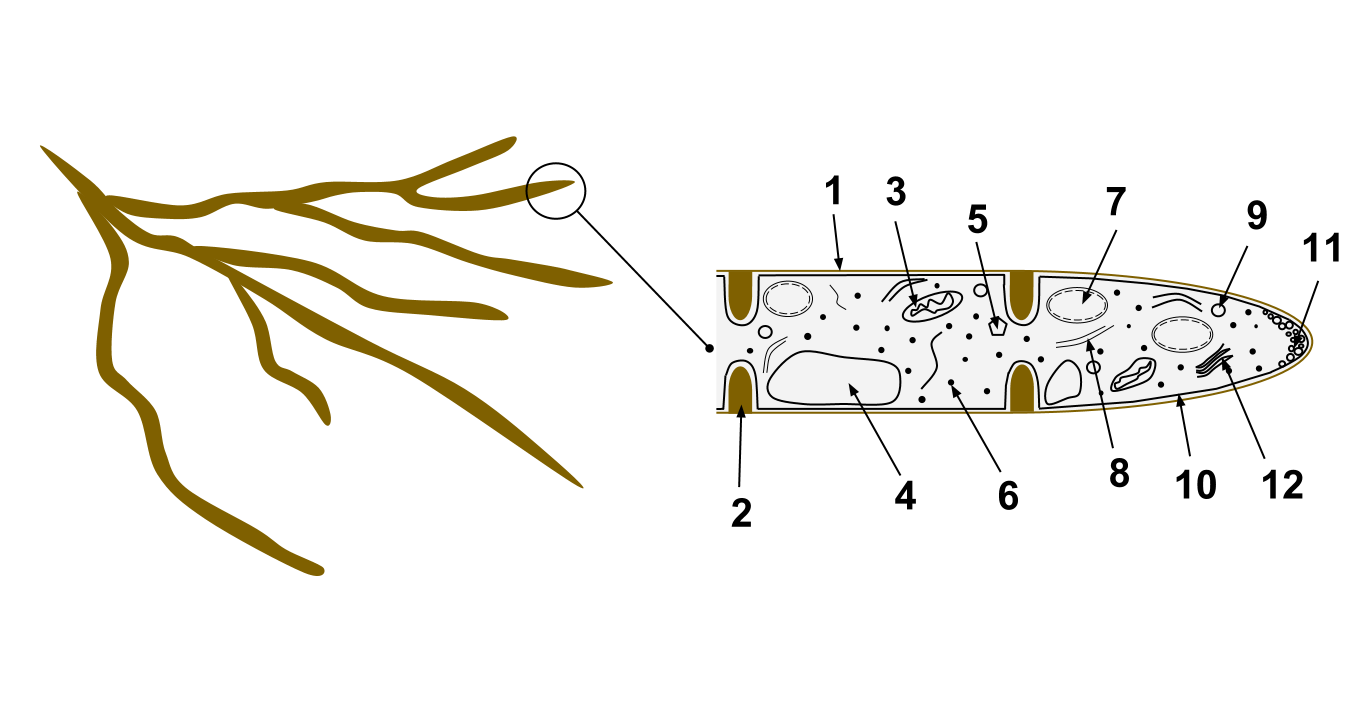 The cells of
The cells of
 In
In
 The origin of the eukaryotic cell, also known as eukaryogenesis, is a milestone in the evolution of life, since eukaryotes include all complex cells and almost all multicellular organisms. A number of approaches have been used to find the first eukaryote and their closest relatives. The last eukaryotic common ancestor (LECA) is the hypothetical last common ancestor of all living eukaryotes, and was most likely a biological population.
Eukaryotes have a number of features that differentiate them from prokaryotes, including an endomembrane system, and unique biochemical pathways such as
The origin of the eukaryotic cell, also known as eukaryogenesis, is a milestone in the evolution of life, since eukaryotes include all complex cells and almost all multicellular organisms. A number of approaches have been used to find the first eukaryote and their closest relatives. The last eukaryotic common ancestor (LECA) is the hypothetical last common ancestor of all living eukaryotes, and was most likely a biological population.
Eukaryotes have a number of features that differentiate them from prokaryotes, including an endomembrane system, and unique biochemical pathways such as
 Alternative proposals include:
* The chronocyte hypothesis postulates that a primitive eukaryotic cell was formed by the endosymbiosis of both archaea and bacteria by a third type of cell, termed a chronocyte. This is mainly to account for the fact that eukaryotic signature proteins were not found anywhere else by 2002.
* The universal common ancestor (UCA) of the current tree of life was a complex organism that survived a mass extinction event rather than an early stage in the evolution of life. Eukaryotes and in particular akaryotes (Bacteria and Archaea) evolved through reductive loss, so that similarities result from differential retention of original features.
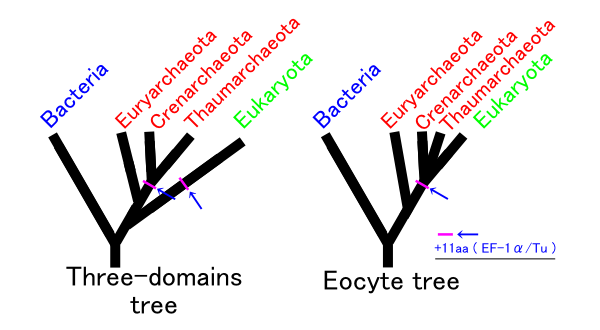
Assuming no other group is involved, there are three possible phylogenies for the Bacteria, Archaea, and Eukaryota in which each is monophyletic. These are labelled 1 to 3 in the table below, with a modification of hypothesis 2 making the 4th column: The ''eocyte hypothesis'', in which the Archaea are paraphyletic. (The table and the names for the hypotheses are based on Harish & Kurland, 2017.)
In recent years, most researchers have favoured either the three domains (3D) or the eocyte hypothesis. An
Alternative proposals include:
* The chronocyte hypothesis postulates that a primitive eukaryotic cell was formed by the endosymbiosis of both archaea and bacteria by a third type of cell, termed a chronocyte. This is mainly to account for the fact that eukaryotic signature proteins were not found anywhere else by 2002.
* The universal common ancestor (UCA) of the current tree of life was a complex organism that survived a mass extinction event rather than an early stage in the evolution of life. Eukaryotes and in particular akaryotes (Bacteria and Archaea) evolved through reductive loss, so that similarities result from differential retention of original features.
Assuming no other group is involved, there are three possible phylogenies for the Bacteria, Archaea, and Eukaryota in which each is monophyletic. These are labelled 1 to 3 in the table below, with a modification of hypothesis 2 making the 4th column: The ''eocyte hypothesis'', in which the Archaea are paraphyletic. (The table and the names for the hypotheses are based on Harish & Kurland, 2017.)
In recent years, most researchers have favoured either the three domains (3D) or the eocyte hypothesis. An
"Eukaryotes"
( Tree of Life Web Project) *
Attraction and sex among our microbial Last Eukaryotic Common Ancestors
The Atlantic, November 11, 2020 {{Authority control Articles containing video clips Eukaryote
organism
In biology, an organism () is any living system that functions as an individual entity. All organisms are composed of cells ( cell theory). Organisms are classified by taxonomy into groups such as multicellular animals, plants, and fu ...
s whose cells
Cell most often refers to:
* Cell (biology), the functional basic unit of life
Cell may also refer to:
Locations
* Monastic cell, a small room, hut, or cave in which a religious recluse lives, alternatively the small precursor of a monastery w ...
have a nucleus
Nucleus ( : nuclei) is a Latin word for the seed inside a fruit. It most often refers to:
* Atomic nucleus, the very dense central region of an atom
*Cell nucleus, a central organelle of a eukaryotic cell, containing most of the cell's DNA
Nucl ...
. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were am ...
and Archaea (both prokaryote
A prokaryote () is a single-celled organism that lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Greek πρό (, 'before') and κάρυον (, 'nut' or 'kernel').Campbell, N. "Biology:Concepts & Con ...
s) make up the other two domains.
The eukaryotes are usually now regarded as having emerged in the Archaea or as a sister of the Asgard archaea. This implies that there are only two domains of life, Bacteria and Archaea, with eukaryotes incorporated among archaea. Eukaryotes represent a small minority of the number of organisms, but, due to their generally much larger size, their collective global biomass
Biomass is plant-based material used as a fuel for heat or electricity production. It can be in the form of wood, wood residues, energy crops, agricultural residues, and waste from industry, farms, and households. Some people use the terms bio ...
is estimated to be about equal to that of prokaryotes. Eukaryotes emerged approximately 2.3–1.8 billion years ago, during the Proterozoic eon, likely as flagellated phagotrophs. Their name comes from the Greek εὖ (''eu'', "well" or "good") and κάρυον (''karyon'', "nut" or "kernel").
Eukaryotic cells typically contain other membrane-bound organelles such as mitochondria
A mitochondrion (; ) is an organelle found in the cells of most Eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used ...
and Golgi apparatus
The Golgi apparatus (), also known as the Golgi complex, Golgi body, or simply the Golgi, is an organelle found in most eukaryotic cells. Part of the endomembrane system in the cytoplasm, it packages proteins into membrane-bound vesicles i ...
. Chloroplast
A chloroplast () is a type of membrane-bound organelle known as a plastid that conducts photosynthesis mostly in plant and algal cells. The photosynthetic pigment chlorophyll captures the energy from sunlight, converts it, and stores it i ...
s can be found in plant
Plants are predominantly photosynthetic eukaryotes of the kingdom Plantae. Historically, the plant kingdom encompassed all living things that were not animals, and included algae and fungi; however, all current definitions of Plantae excl ...
s and algae
Algae (; singular alga ) is an informal term for a large and diverse group of photosynthetic eukaryotic organisms. It is a polyphyletic grouping that includes species from multiple distinct clades. Included organisms range from unicellular micr ...
. Prokaryotic cells may contain primitive organelles. Eukaryotes may be either unicellular or multicellular
A multicellular organism is an organism that consists of more than one cell, in contrast to unicellular organism.
All species of animals, land plants and most fungi are multicellular, as are many algae, whereas a few organisms are partially ...
, and include many cell type
A cell type is a classification used to identify cells that share morphological or phenotypical features. A multicellular organism may contain cells of a number of widely differing and specialized cell types, such as muscle cells and skin cell ...
s forming different kinds of tissue. In comparison, prokaryotes are typically unicellular. Animal
Animals are multicellular, eukaryotic organisms in the biological kingdom Animalia. With few exceptions, animals consume organic material, breathe oxygen, are able to move, can reproduce sexually, and go through an ontogenetic stage ...
s, plant
Plants are predominantly photosynthetic eukaryotes of the kingdom Plantae. Historically, the plant kingdom encompassed all living things that were not animals, and included algae and fungi; however, all current definitions of Plantae excl ...
s, and fungi
A fungus ( : fungi or funguses) is any member of the group of eukaryotic organisms that includes microorganisms such as yeasts and molds, as well as the more familiar mushrooms. These organisms are classified as a kingdom, separately fr ...
are the most familiar eukaryotes. Other eukaryotes are sometimes called protist
A protist () is any eukaryotic organism (that is, an organism whose cells contain a cell nucleus) that is not an animal, plant, or fungus. While it is likely that protists share a common ancestor (the last eukaryotic common ancestor), the e ...
s.
Eukaryotes can reproduce both asexually through mitosis and sexually through meiosis
Meiosis (; , since it is a reductional division) is a special type of cell division of germ cells in sexually-reproducing organisms that produces the gametes, such as sperm or egg cells. It involves two rounds of division that ultimately ...
and gamete
A gamete (; , ultimately ) is a haploid cell that fuses with another haploid cell during fertilization in organisms that reproduce sexually. Gametes are an organism's reproductive cells, also referred to as sex cells. In species that produce ...
fusion. In mitosis, one cell divides to produce two genetically identical cells. In meiosis, DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritan ...
is followed by two rounds of cell division
Cell division is the process by which a parent cell divides into two daughter cells. Cell division usually occurs as part of a larger cell cycle in which the cell grows and replicates its chromosome(s) before dividing. In eukaryotes, there ...
to produce four haploid daughter cells that act as sex cells or gametes. Each gamete has just one set of chromosomes, each a unique mix of the corresponding pair of parental chromosome
A chromosome is a long DNA molecule with part or all of the genetic material of an organism. In most chromosomes the very long thin DNA fibers are coated with packaging proteins; in eukaryotic cells the most important of these proteins ar ...
s resulting from genetic recombination during meiosis.
Cell features
Eukaryotic cells
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bact ...
are typically much larger than those of prokaryote
A prokaryote () is a single-celled organism that lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Greek πρό (, 'before') and κάρυον (, 'nut' or 'kernel').Campbell, N. "Biology:Concepts & Con ...
s, having a volume of around 10,000 times greater than the prokaryotic cell. They have a variety of internal membrane-bound structures, called organelles, and a cytoskeleton
The cytoskeleton is a complex, dynamic network of interlinking protein filaments present in the cytoplasm of all cells, including those of bacteria and archaea. In eukaryotes, it extends from the cell nucleus to the cell membrane and is co ...
composed of microtubules, microfilament
Microfilaments, also called actin filaments, are protein filaments in the cytoplasm of eukaryotic cells that form part of the cytoskeleton. They are primarily composed of polymers of actin, but are modified by and interact with numerous other pr ...
s, and intermediate filaments, which play an important role in defining the cell's organization and shape. Eukaryotic DNA is divided into several linear bundles called chromosome
A chromosome is a long DNA molecule with part or all of the genetic material of an organism. In most chromosomes the very long thin DNA fibers are coated with packaging proteins; in eukaryotic cells the most important of these proteins ar ...
s, which are separated by a microtubular spindle during nuclear division.
Internal membranes
vesicles
Vesicle may refer to:
; In cellular biology or chemistry
* Vesicle (biology and chemistry), a supramolecular assembly of lipid molecules, like a cell membrane
* Synaptic vesicle
; In human embryology
* Vesicle (embryology), bulge-like features o ...
and vacuole
A vacuole () is a membrane-bound organelle which is present in plant and fungal cells and some protist, animal, and bacterial cells. Vacuoles are essentially enclosed compartments which are filled with water containing inorganic and organic m ...
s, can form by budding off other membranes. Many cells ingest food and other materials through a process of endocytosis, where the outer membrane invaginates and then pinches off to form a vesicle. It is probable that most other membrane-bound organelles are ultimately derived from such vesicles. Alternatively some products produced by the cell can leave in a vesicle through exocytosis.
The nucleus is surrounded by a double membrane known as the nuclear envelope, with nuclear pore
A nuclear pore is a part of a large complex of proteins, known as a nuclear pore complex that spans the nuclear envelope, which is the double membrane surrounding the eukaryotic cell nucleus. There are approximately 1,000 nuclear pore comple ...
s that allow material to move in and out. Various tube- and sheet-like extensions of the nuclear membrane form the endoplasmic reticulum
The endoplasmic reticulum (ER) is, in essence, the transportation system of the eukaryotic cell, and has many other important functions such as protein folding. It is a type of organelle made up of two subunits – rough endoplasmic reticulum ...
, which is involved in protein transport and maturation. It includes the rough endoplasmic reticulum where ribosomes are attached to synthesize proteins, which enter the interior space or lumen. Subsequently, they generally enter vesicles, which bud off from the smooth endoplasmic reticulum. In most eukaryotes, these protein-carrying vesicles are released and further modified in stacks of flattened vesicles (cisterna
A cisterna (plural cisternae) is a flattened membrane vesicle found in the endoplasmic reticulum and Golgi apparatus. Cisternae are an integral part of the packaging and modification processes of proteins occurring in the Golgi.
Function
Protei ...
e), the Golgi apparatus
The Golgi apparatus (), also known as the Golgi complex, Golgi body, or simply the Golgi, is an organelle found in most eukaryotic cells. Part of the endomembrane system in the cytoplasm, it packages proteins into membrane-bound vesicles i ...
.
Vesicles may be specialized for various purposes. For instance, lysosome
A lysosome () is a membrane-bound organelle found in many animal cells. They are spherical vesicles that contain hydrolytic enzymes that can break down many kinds of biomolecules. A lysosome has a specific composition, of both its membrane p ...
s contain digestive enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products ...
s that break down most biomolecules in the cytoplasm. Peroxisomes are used to break down peroxide
In chemistry, peroxides are a group of compounds with the structure , where R = any element. The group in a peroxide is called the peroxide group or peroxo group. The nomenclature is somewhat variable.
The most common peroxide is hydrogen p ...
, which is otherwise toxic. Many protozoans have contractile vacuoles, which collect and expel excess water, and extrusomes, which expel material used to deflect predators or capture prey. In higher plants, most of a cell's volume is taken up by a central vacuole, which mostly contains water and primarily maintains its osmotic pressure.
Mitochondria
Mitochondria
A mitochondrion (; ) is an organelle found in the cells of most Eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used ...
are organelles found in all but one eukaryote, and are commonly referred to as "the powerhouse of the cell". Mitochondria provide energy to the eukaryote cell by oxidising sugars or fats and releasing energy as ATP. They have two surrounding membranes
A membrane is a selective barrier; it allows some things to pass through but stops others. Such things may be molecules, ions, or other small particles. Membranes can be generally classified into synthetic membranes and biological membranes. Bi ...
, each a phospholipid bi-layer; the inner
Interior may refer to:
Arts and media
* ''Interior'' (Degas) (also known as ''The Rape''), painting by Edgar Degas
* ''Interior'' (play), 1895 play by Belgian playwright Maurice Maeterlinck
* ''The Interior'' (novel), by Lisa See
* Interior de ...
of which is folded into invaginations called cristae where aerobic respiration
Cellular respiration is the process by which biological fuels are oxidised in the presence of an inorganic electron acceptor such as oxygen to produce large amounts of energy, to drive the bulk production of ATP. Cellular respiration may be des ...
takes place.
The outer mitochondrial membrane is freely permeable and allows almost anything to enter into the intermembrane space while the inner mitochondrial membrane is semi permeable so allows only some required things into the mitochondrial matrix.
Mitochondria contain their own DNA, which has close structural similarities to bacterial DNA, and which encodes rRNA
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribos ...
and tRNA
Transfer RNA (abbreviated tRNA and formerly referred to as sRNA, for soluble RNA) is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes), that serves as the physical link between the mRNA and the amino ...
genes that produce RNA which is closer in structure to bacterial RNA than to eukaryote RNA. They are now generally held to have developed from endosymbiotic prokaryotes, probably Alphaproteobacteria.
Some eukaryotes, such as the metamonads such as '' Giardia'' and ''Trichomonas
''Trichomonas'' is a genus of anaerobic excavate parasites of vertebrates. It was first discovered by Alfred François Donné in 1836 when he found these parasites in the pus of a patient suffering from vaginitis, an inflammation of the vagina ...
'', and the amoebozoan '' Pelomyxa'', appear to lack mitochondria, but all have been found to contain mitochondrion-derived organelles, such as hydrogenosomes and mitosomes, and thus have lost their mitochondria secondarily. They obtain energy by enzymatic action on nutrients absorbed from the environment. The metamonad ''Monocercomonoides
''Monocercomonoides'' is a genus of flagellate Excavata belonging to the order Oxymonadida. It was established by Bernard V. Travis and was first described as those with "polymastiginid flagellates having three anterior flagella and a trailing ...
'' has also acquired, by lateral gene transfer, a cytosolic sulfur
Sulfur (or sulphur in British English) is a chemical element with the symbol S and atomic number 16. It is abundant, multivalent and nonmetallic. Under normal conditions, sulfur atoms form cyclic octatomic molecules with a chemical formul ...
mobilisation system which provides the clusters of iron and sulfur required for protein synthesis. The normal mitochondrial iron-sulfur cluster pathway has been lost secondarily.
Plastids
Plants and various groups ofalgae
Algae (; singular alga ) is an informal term for a large and diverse group of photosynthetic eukaryotic organisms. It is a polyphyletic grouping that includes species from multiple distinct clades. Included organisms range from unicellular micr ...
also have plastids. Plastids also have their own DNA and are developed from endosymbionts, in this case cyanobacteria
Cyanobacteria (), also known as Cyanophyta, are a phylum of gram-negative bacteria that obtain energy via photosynthesis. The name ''cyanobacteria'' refers to their color (), which similarly forms the basis of cyanobacteria's common name, bl ...
. They usually take the form of chloroplast
A chloroplast () is a type of membrane-bound organelle known as a plastid that conducts photosynthesis mostly in plant and algal cells. The photosynthetic pigment chlorophyll captures the energy from sunlight, converts it, and stores it i ...
s which, like cyanobacteria, contain chlorophyll and produce organic compounds (such as glucose
Glucose is a simple sugar with the molecular formula . Glucose is overall the most abundant monosaccharide, a subcategory of carbohydrates. Glucose is mainly made by plants and most algae during photosynthesis from water and carbon dioxide, u ...
) through photosynthesis
Photosynthesis is a process used by plants and other organisms to convert light energy into chemical energy that, through cellular respiration, can later be released to fuel the organism's activities. Some of this chemical energy is stored in ...
. Others are involved in storing food. Although plastids probably had a single origin, not all plastid-containing groups are closely related. Instead, some eukaryotes have obtained them from others through secondary endosymbiosis or ingestion. The capture and sequestering of photosynthetic cells and chloroplasts occurs in many types of modern eukaryotic organisms and is known as kleptoplasty.
Endosymbiotic origins have also been proposed for the nucleus, and for eukaryotic flagella.
Cytoskeletal structures
 Many eukaryotes have long slender motile cytoplasmic projections, called flagella, or similar structures called cilia. Flagella and cilia are sometimes referred to as
Many eukaryotes have long slender motile cytoplasmic projections, called flagella, or similar structures called cilia. Flagella and cilia are sometimes referred to as undulipodia
An undulipodium or undulopodium (a Greek word meaning "swinging foot"), or a 9+2 organelle is a motile filamentous extracellular projection of eukaryotic cells. It is basically synonymous to flagella and cilia which are differing terms for simila ...
, and are variously involved in movement, feeding, and sensation. They are composed mainly of tubulin. These are entirely distinct from prokaryotic flagellae. They are supported by a bundle of microtubules arising from a centriole
In cell biology a centriole is a cylindrical organelle composed mainly of a protein called tubulin. Centrioles are found in most eukaryotic cells, but are not present in conifers (Pinophyta), flowering plants (angiosperms) and most fungi, and are ...
, characteristically arranged as nine doublets surrounding two singlets. Flagella also may have hairs, or mastigonemes, and scales connecting membranes and internal rods. Their interior is continuous with the cell's cytoplasm
In cell biology, the cytoplasm is all of the material within a eukaryotic cell, enclosed by the cell membrane, except for the cell nucleus. The material inside the nucleus and contained within the nuclear membrane is termed the nucleoplasm. ...
.
Microfilamental structures composed of actin
Actin is a family of globular multi-functional proteins that form microfilaments in the cytoskeleton, and the thin filaments in muscle fibrils. It is found in essentially all eukaryotic cells, where it may be present at a concentration of ov ...
and actin binding proteins, e.g., α- actinin, fimbrin, filamin are present in submembranous cortical layers and bundles, as well. Motor proteins of microtubules, e.g., dynein or kinesin and actin, e.g., myosins
Myosins () are a superfamily of motor proteins best known for their roles in muscle contraction and in a wide range of other motility processes in eukaryotes. They are ATP-dependent and responsible for actin-based motility.
The first myosin (M ...
provide dynamic character of the network.
Centrioles are often present even in cells and groups that do not have flagella, but conifers and flowering plant
Flowering plants are plants that bear flowers and fruits, and form the clade Angiospermae (), commonly called angiosperms. The term "angiosperm" is derived from the Greek words ('container, vessel') and ('seed'), and refers to those plants t ...
s have neither. They generally occur in groups that give rise to various microtubular roots. These form a primary component of the cytoskeletal structure, and are often assembled over the course of several cell divisions, with one flagellum retained from the parent and the other derived from it. Centrioles produce the spindle during nuclear division.
The significance of cytoskeletal structures is underlined in the determination of shape of the cells, as well as their being essential components of migratory responses like chemotaxis and chemokinesis. Some protists have various other microtubule-supported organelles. These include the radiolaria and heliozoa
Heliozoa, commonly known as sun-animalcules, are microbial eukaryotes ( protists) with stiff arms (axopodia) radiating from their spherical bodies, which are responsible for their common name. The axopodia are microtubule-supported projections fr ...
, which produce axopodia used in flotation or to capture prey, and the haptophytes, which have a peculiar flagellum-like organelle called the haptonema.
Cell wall
The cells of plants and algae, fungi and mostchromalveolates
Chromista is a biological kingdom consisting of single-celled and multicellular eukaryotic species that share similar features in their photosynthetic organelles (plastids). It includes all protists whose plastids contain chlorophyll ''c'', such ...
have a cell wall, a layer outside the cell membrane
The cell membrane (also known as the plasma membrane (PM) or cytoplasmic membrane, and historically referred to as the plasmalemma) is a biological membrane that separates and protects the interior of all cells from the outside environment (t ...
, providing the cell with structural support, protection, and a filtering mechanism. The cell wall also prevents over-expansion when water enters the cell.
The major polysaccharides making up the primary cell wall of land plants
The Embryophyta (), or land plants, are the most familiar group of green plants that comprise vegetation on Earth. Embryophytes () have a common ancestor with green algae, having emerged within the Phragmoplastophyta clade of green algae as siste ...
are cellulose
Cellulose is an organic compound with the formula , a polysaccharide consisting of a linear chain of several hundred to many thousands of β(1→4) linked D-glucose units. Cellulose is an important structural component of the primary cell wa ...
, hemicellulose, and pectin
Pectin ( grc, πηκτικός ': "congealed" and "curdled") is a heteropolysaccharide, a structural acid contained in the primary lamella, in the middle lamella, and in the cell walls of terrestrial plants. The principal, chemical component o ...
. The cellulose microfibrils are linked via hemicellulosic tethers to form the cellulose-hemicellulose network, which is embedded in the pectin matrix. The most common hemicellulose in the primary cell wall is xyloglucan.
Differences among eukaryotic cells
There are many different types of eukaryotic cells, though animals and plants are the most familiar eukaryotes, and thus provide an excellent starting point for understanding eukaryotic structure. Fungi and many protists have some substantial differences, however.Animal cell
cell wall
A cell wall is a structural layer surrounding some types of cells, just outside the cell membrane. It can be tough, flexible, and sometimes rigid. It provides the cell with both structural support and protection, and also acts as a filtering mec ...
s and chloroplast
A chloroplast () is a type of membrane-bound organelle known as a plastid that conducts photosynthesis mostly in plant and algal cells. The photosynthetic pigment chlorophyll captures the energy from sunlight, converts it, and stores it i ...
s and have smaller vacuole
A vacuole () is a membrane-bound organelle which is present in plant and fungal cells and some protist, animal, and bacterial cells. Vacuoles are essentially enclosed compartments which are filled with water containing inorganic and organic m ...
s. Due to the lack of a cell wall
A cell wall is a structural layer surrounding some types of cells, just outside the cell membrane. It can be tough, flexible, and sometimes rigid. It provides the cell with both structural support and protection, and also acts as a filtering mec ...
, animal cells can transform into a variety of shapes. A phagocytic cell can even engulf other structures.
Plant cell
Plant cell
Plant cells are the cells present in green plants, photosynthetic eukaryotes of the kingdom Plantae. Their distinctive features include primary cell walls containing cellulose, hemicelluloses and pectin, the presence of plastids with the capab ...
s have a number of features that distinguish them from the cells of the other eukaryotic organisms. These include:
* A large central vacuole
A vacuole () is a membrane-bound organelle which is present in plant and fungal cells and some protist, animal, and bacterial cells. Vacuoles are essentially enclosed compartments which are filled with water containing inorganic and organic ...
(enclosed by a membrane, the tonoplast), which maintains the cell's turgor and controls movement of molecule
A molecule is a group of two or more atoms held together by attractive forces known as chemical bonds; depending on context, the term may or may not include ions which satisfy this criterion. In quantum physics, organic chemistry, and b ...
s between the cytosol and sap
* A primary cell wall
A cell wall is a structural layer surrounding some types of cells, just outside the cell membrane. It can be tough, flexible, and sometimes rigid. It provides the cell with both structural support and protection, and also acts as a filtering mec ...
containing cellulose
Cellulose is an organic compound with the formula , a polysaccharide consisting of a linear chain of several hundred to many thousands of β(1→4) linked D-glucose units. Cellulose is an important structural component of the primary cell wa ...
, hemicellulose and pectin
Pectin ( grc, πηκτικός ': "congealed" and "curdled") is a heteropolysaccharide, a structural acid contained in the primary lamella, in the middle lamella, and in the cell walls of terrestrial plants. The principal, chemical component o ...
, deposited by the protoplast
Protoplast (), is a biological term coined by Hanstein in 1880 to refer to the entire cell, excluding the cell wall. Protoplasts can be generated by stripping the cell wall from plant, bacterial, or fungal cells by mechanical, chemical or e ...
on the outside of the cell membrane
The cell membrane (also known as the plasma membrane (PM) or cytoplasmic membrane, and historically referred to as the plasmalemma) is a biological membrane that separates and protects the interior of all cells from the outside environment (t ...
; this contrasts with the cell walls of fungi
A fungus ( : fungi or funguses) is any member of the group of eukaryotic organisms that includes microorganisms such as yeasts and molds, as well as the more familiar mushrooms. These organisms are classified as a kingdom, separately fr ...
, which contain chitin, and the cell envelopes of prokaryotes, in which peptidoglycans are the main structural molecules
* The plasmodesmata, pores in the cell wall that link adjacent cells and allow plant cells to communicate with adjacent cells. Animals have a different but functionally analogous system of gap junction
Gap junctions are specialized intercellular connections between a multitude of animal cell-types. They directly connect the cytoplasm of two cells, which allows various molecules, ions and electrical impulses to directly pass through a regula ...
s between adjacent cells.
* Plastids, especially chloroplast
A chloroplast () is a type of membrane-bound organelle known as a plastid that conducts photosynthesis mostly in plant and algal cells. The photosynthetic pigment chlorophyll captures the energy from sunlight, converts it, and stores it i ...
s, organelles that contain chlorophyll, the pigment that gives plant
Plants are predominantly photosynthetic eukaryotes of the kingdom Plantae. Historically, the plant kingdom encompassed all living things that were not animals, and included algae and fungi; however, all current definitions of Plantae excl ...
s their green color and allows them to perform photosynthesis
Photosynthesis is a process used by plants and other organisms to convert light energy into chemical energy that, through cellular respiration, can later be released to fuel the organism's activities. Some of this chemical energy is stored in ...
* Bryophytes and seedless vascular plants only have flagellae and centrioles in the sperm cells. Sperm of cycad
Cycads are seed plants that typically have a stout and woody (ligneous) trunk with a crown of large, hard, stiff, evergreen and (usually) pinnate leaves. The species are dioecious, that is, individual plants of a species are either male o ...
s and '' Ginkgo'' are large, complex cells that swim with hundreds to thousands of flagellae.
* Conifers (Pinophyta) and flowering plant
Flowering plants are plants that bear flowers and fruits, and form the clade Angiospermae (), commonly called angiosperms. The term "angiosperm" is derived from the Greek words ('container, vessel') and ('seed'), and refers to those plants t ...
s (Angiospermae) lack the flagellae and centriole
In cell biology a centriole is a cylindrical organelle composed mainly of a protein called tubulin. Centrioles are found in most eukaryotic cells, but are not present in conifers (Pinophyta), flowering plants (angiosperms) and most fungi, and are ...
s that are present in animal cells.
Fungal cell
 The cells of
The cells of fungi
A fungus ( : fungi or funguses) is any member of the group of eukaryotic organisms that includes microorganisms such as yeasts and molds, as well as the more familiar mushrooms. These organisms are classified as a kingdom, separately fr ...
are similar to animal cells, with the following exceptions:
* A cell wall that contains chitin
* Less compartmentation between cells; the hyphae of higher fungi have porous partitions called septa
The Southeastern Pennsylvania Transportation Authority (SEPTA) is a regional public transportation authority that operates bus, rapid transit, commuter rail, light rail, and electric trolleybus services for nearly 4 million people in five c ...
, which allow the passage of cytoplasm, organelles, and, sometimes, nuclei; so each organism is essentially a giant multinucleate supercell – these fungi are described as coenocytic. Primitive fungi have few or no septa.
* Only the most primitive fungi, chytrids, have flagella.
Other eukaryotic cells
Some groups of eukaryotes have unique organelles, such as the cyanelles (unusual plastids) of the glaucophytes, the haptonema of the haptophytes, or the ejectosomes of the cryptomonads. Other structures, such as pseudopodia, are found in various eukaryote groups in different forms, such as the lobose amoebozoans or the reticulose foraminiferans. Structures known as cortical alveoli are vesicles present under the cell membrane of manyprotist
A protist () is any eukaryotic organism (that is, an organism whose cells contain a cell nucleus) that is not an animal, plant, or fungus. While it is likely that protists share a common ancestor (the last eukaryotic common ancestor), the e ...
s such as dinoflagellates, ciliate
The ciliates are a group of alveolates characterized by the presence of hair-like organelles called cilia, which are identical in structure to eukaryotic flagella, but are in general shorter and present in much larger numbers, with a differen ...
s and apicomplexan parasites.
Reproduction

Cell division
Cell division is the process by which a parent cell divides into two daughter cells. Cell division usually occurs as part of a larger cell cycle in which the cell grows and replicates its chromosome(s) before dividing. In eukaryotes, there ...
generally takes place asexually by mitosis, a process that allows each daughter nucleus to receive one copy of each chromosome
A chromosome is a long DNA molecule with part or all of the genetic material of an organism. In most chromosomes the very long thin DNA fibers are coated with packaging proteins; in eukaryotic cells the most important of these proteins ar ...
. Most eukaryotes also have a life cycle that involves sexual reproduction
Sexual reproduction is a type of reproduction that involves a complex life cycle in which a gamete ( haploid reproductive cells, such as a sperm or egg cell) with a single set of chromosomes combines with another gamete to produce a zygote th ...
, alternating between a haploid phase, where only one copy of each chromosome is present in each cell and a diploid
Ploidy () is the number of complete sets of chromosomes in a cell, and hence the number of possible alleles for autosomal and pseudoautosomal genes. Sets of chromosomes refer to the number of maternal and paternal chromosome copies, respectiv ...
phase, wherein two copies of each chromosome are present in each cell. The diploid phase is formed by fusion of two haploid gametes to form a zygote, which may divide by mitosis or undergo chromosome reduction by meiosis
Meiosis (; , since it is a reductional division) is a special type of cell division of germ cells in sexually-reproducing organisms that produces the gametes, such as sperm or egg cells. It involves two rounds of division that ultimately ...
. There is considerable variation in this pattern. Animals have no multicellular haploid phase, but each plant generation can consist of haploid and diploid multicellular phases.
Eukaryotes have a smaller surface area to volume ratio than prokaryotes, and thus have lower metabolic rates and longer generation times.
The evolution of sexual reproduction may be a primordial and fundamental characteristic of eukaryotes. Based on a phylogenetic analysis, Dacks and Roger
Roger is a given name, usually masculine, and a surname. The given name is derived from the Old French personal names ' and '. These names are of Germanic origin, derived from the elements ', ''χrōþi'' ("fame", "renown", "honour") and ', ' ( ...
proposed that facultative sex was present in the common ancestor of all eukaryotes. A core set of genes that function in meiosis is present in both ''Trichomonas vaginalis
''Trichomonas vaginalis'' is an anaerobic, flagellated protozoan parasite and the causative agent of a sexually transmitted disease called trichomoniasis. It is the most common pathogenic protozoan that infects humans in industrialized countri ...
'' and '' Giardia intestinalis'', two organisms previously thought to be asexual. Since these two species are descendants of lineages that diverged early from the eukaryotic evolutionary tree, it was inferred that core meiotic genes, and hence sex, were likely present in a common ancestor of all eukaryotes. Eukaryotic species once thought to be asexual, such as parasitic protozoa of the genus '' Leishmania'', have been shown to have a sexual cycle. Also, evidence now indicates that amoebae, previously regarded as asexual, are anciently sexual and that the majority of present-day asexual groups likely arose recently and independently.
Classification

 In
In antiquity
Antiquity or Antiquities may refer to:
Historical objects or periods Artifacts
*Antiquities, objects or artifacts surviving from ancient cultures
Eras
Any period before the European Middle Ages (5th to 15th centuries) but still within the histo ...
, the two lineages of animal
Animals are multicellular, eukaryotic organisms in the biological kingdom Animalia. With few exceptions, animals consume organic material, breathe oxygen, are able to move, can reproduce sexually, and go through an ontogenetic stage ...
s and plant
Plants are predominantly photosynthetic eukaryotes of the kingdom Plantae. Historically, the plant kingdom encompassed all living things that were not animals, and included algae and fungi; however, all current definitions of Plantae excl ...
s were recognized. They were given the taxonomic rank
In biological classification, taxonomic rank is the relative level of a group of organisms (a taxon) in an ancestral or hereditary hierarchy. A common system consists of species, genus, family, order, class, phylum, kingdom, domain. While ...
of Kingdom by Linnaeus. Though he included the fungi
A fungus ( : fungi or funguses) is any member of the group of eukaryotic organisms that includes microorganisms such as yeasts and molds, as well as the more familiar mushrooms. These organisms are classified as a kingdom, separately fr ...
with plants with some reservations, it was later realized that they are quite distinct and warrant a separate kingdom, the composition of which was not entirely clear until the 1980s. The various single-cell eukaryotes were originally placed with plants or animals when they became known. In 1818, the German biologist Georg A. Goldfuss
Georg August Goldfuss (Goldfuß, 18 April 1782 – 2 October 1848) was a German palaeontologist, zoologist and botanist.
Goldfuss was born at Thurnau near Bayreuth. He was educated at Erlangen, where he graduated PhD in 1804 and became pro ...
coined the word '' protozoa'' to refer to organisms such as ciliate
The ciliates are a group of alveolates characterized by the presence of hair-like organelles called cilia, which are identical in structure to eukaryotic flagella, but are in general shorter and present in much larger numbers, with a differen ...
s, and this group was expanded until it encompassed all single-celled eukaryotes, and given their own kingdom, the Protista
A protist () is any eukaryotic organism (that is, an organism whose cells contain a cell nucleus) that is not an animal, plant, or fungus. While it is likely that protists share a common ancestor (the last eukaryotic common ancestor), the e ...
, by Ernst Haeckel
Ernst Heinrich Philipp August Haeckel (; 16 February 1834 – 9 August 1919) was a German zoologist, naturalist, eugenicist, philosopher, physician, professor, marine biologist and artist. He discovered, described and named thousands of new s ...
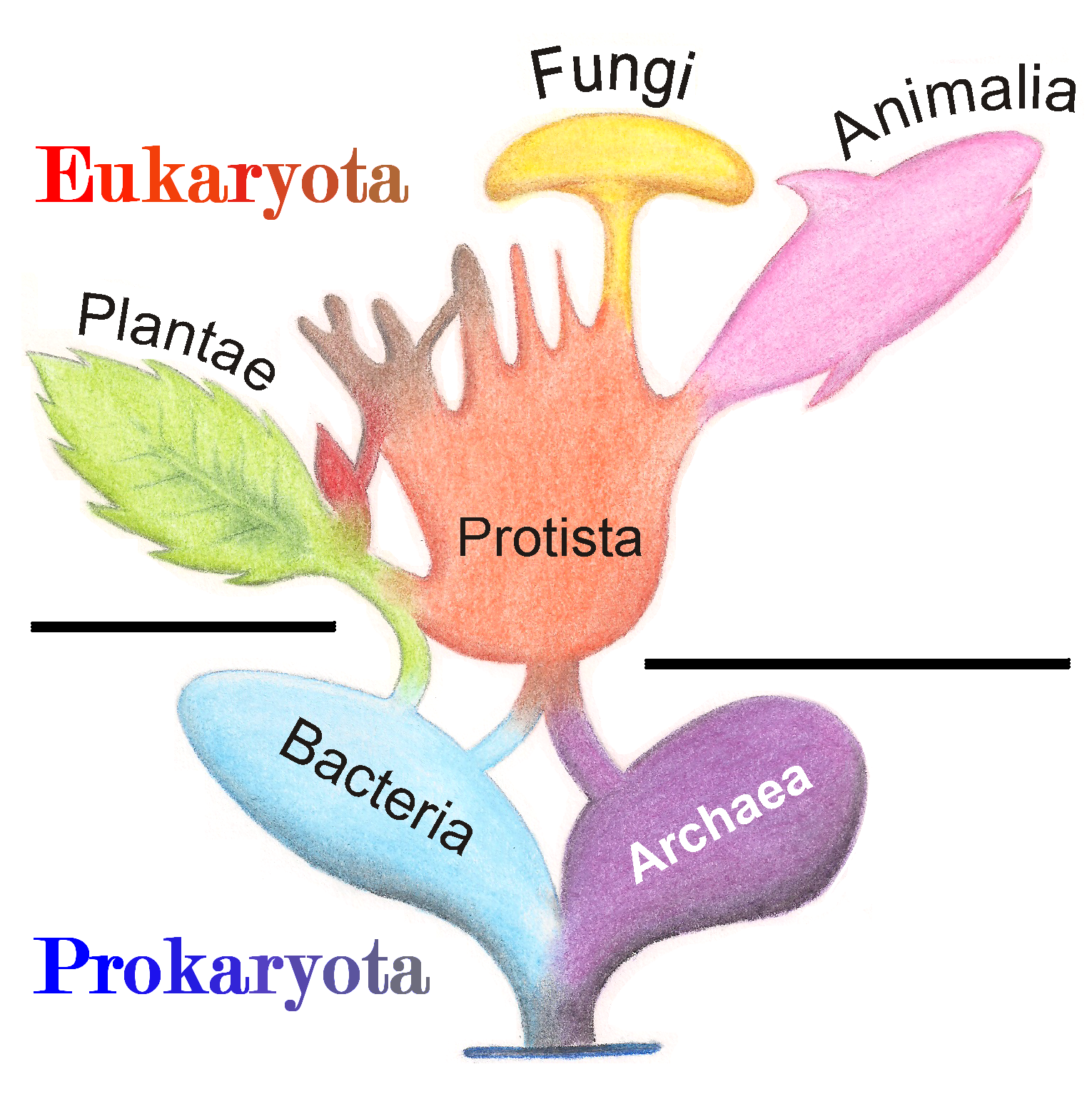
in 1866. The eukaryotes thus came to be composed of four kingdoms:
* Kingdom Protista
A protist () is any eukaryotic organism (that is, an organism whose cells contain a cell nucleus) that is not an animal, plant, or fungus. While it is likely that protists share a common ancestor (the last eukaryotic common ancestor), the e ...
* Kingdom Plantae
* Kingdom Fungi
A fungus ( : fungi or funguses) is any member of the group of eukaryotic organisms that includes microorganisms such as yeasts and molds, as well as the more familiar mushrooms. These organisms are classified as a kingdom, separately fr ...
* Kingdom Animalia
Animals are multicellular, eukaryotic organisms in the biological kingdom Animalia. With few exceptions, animals consume organic material, breathe oxygen, are able to move, can reproduce sexually, and go through an ontogenetic stage in ...
The protists were understood to be "primitive forms", and thus an evolutionary grade, united by their primitive unicellular nature. The disentanglement of the deep splits in the tree of life
The tree of life is a fundamental archetype in many of the world's mythological, religious, and philosophical traditions. It is closely related to the concept of the sacred tree.Giovino, Mariana (2007). ''The Assyrian Sacred Tree: A Hist ...
only really started with DNA sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. T ...
, leading to a system of domains rather than kingdoms as top level rank being put forward by Carl Woese, uniting all the eukaryote kingdoms under the eukaryote domain. At the same time, work on the protist tree intensified, and is still actively going on today. Several alternative classifications have been forwarded, though there is no consensus in the field.
Eukaryotes are a clade usually assessed to be sister to Heimdallarchaeota
Asgard or Asgardarchaeota is a proposed superphylum consisting of a group of archaea that includes Lokiarchaeota, Thorarchaeota, Odinarchaeota, and Heimdallarchaeota. It appears the eukaryotes emerged within the Asgard, in a branch containing ...
in the Asgard grouping in the Archaea. In one proposed system, the basal groupings are the Opimoda, Diphoda
A bikont ("two flagella") is any of the eukaryotic organisms classified in the group Bikonta. Many single-celled members of the group, and the presumed ancestor, have two flagella.
Enzymes
Another shared trait of bikonts is the fusion of two ...
, the Discoba
Excavata is a major supergroup of unicellular organisms belonging to the domain Eukaryota. It was first suggested by Simpson and Patterson in 1999 and introduced by Thomas Cavalier-Smith in 2002 as a formal taxon. It contains a variety of fr ...
, and the Loukozoa. The Eukaryote root is usually assessed to be near or even in Discoba.
A classification produced in 2005 for the International Society of Protistologists, which reflected the consensus of the time, divided the eukaryotes into six supposedly monophyletic ' supergroups'. However, in the same year (2005), doubts were expressed as to whether some of these supergroups were monophyletic, particularly the Chromalveolata, and a review in 2006 noted the lack of evidence for several of the supposed six supergroups. A revised classification in 2012 recognizes five supergroups.
There are also smaller groups of eukaryotes whose position is uncertain or seems to fall outside the major groups – in particular, Haptophyta, Cryptophyta, Centrohelida, Telonemia, Picozoa, Apusomonadida, Ancyromonadida, Breviatea, and the genus ''Collodictyon
''Collodictyon'' is a genus of single-celled, omnivorous eukaryotes belonging to the collodictyonids, also known as diphylleids. Due to their mix of cellular components, Collodictyonids do not belong to any well-known kingdom-level grouping of th ...
''. Overall, it seems that, although progress has been made, there are still very significant uncertainties in the evolutionary history and classification of eukaryotes. As Roger
Roger is a given name, usually masculine, and a surname. The given name is derived from the Old French personal names ' and '. These names are of Germanic origin, derived from the elements ', ''χrōþi'' ("fame", "renown", "honour") and ', ' ( ...
& Simpson said in 2009 "with the current pace of change in our understanding of the eukaryote tree of life, we should proceed with caution." Newly identified protists, purported to represent novel, deep-branching lineages, continue to be described well into the 21st century; recent examples including ''Rhodelphis
''Rhodelphis'' is a single-celled archaeplastid that lives in aquatic environments and is the sister group to red algae and possibly Picozoa. While red algae have no flagellated stages and are generally photoautotrophic, ''Rhodelphis'' is a flage ...
'', putative sister group to Rhodophyta, and '' Anaeramoeba'', anaerobic amoebaflagellates of uncertain placement.
Phylogeny
TherRNA
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribos ...
trees constructed during the 1980s and 1990s left most eukaryotes in an unresolved "crown" group (not technically a true crown), which was usually divided by the form of the mitochondrial cristae; see crown eukaryotes
Taking into account the definition of a crown group, crown eukaryotes could be seen as the aggrupation of all lineages descending from LECA (Last Eukarytotic Common Ancestor). This comprises a huge ensemble of taxa that seemingly diverged simult ...
. The few groups that lack mitochondria
A mitochondrion (; ) is an organelle found in the cells of most Eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used ...
branched separately, and so the absence was believed to be primitive; but this is now considered an artifact of long-branch attraction
In phylogenetics, long branch attraction (LBA) is a form of systematic error whereby distantly related lineages are incorrectly inferred to be closely related. LBA arises when the amount of molecular or morphological change accumulated within a lin ...
, and they are known to have lost them secondarily.
It has been estimated that there may be 75 distinct lineages of eukaryotes. Most of these lineages are protists.
The known eukaryote genome sizes vary from 8.2 megabases (Mb) in '' Babesia bovis'' to 112,000–220,050 Mb in the dinoflagellate ''Prorocentrum micans
The Prorocentrales are a small order of dinoflagellates. They are distinguished by having their two flagella inserted apically, rather than ventrally as in other groups. One flagellum extends forward and the other circles its base, and there are ...
'', showing that the genome of the ancestral eukaryote has undergone considerable variation during its evolution. The last common ancestor of all eukaryotes is believed to have been a phagotrophic protist with a nucleus, at least one centriole
In cell biology a centriole is a cylindrical organelle composed mainly of a protein called tubulin. Centrioles are found in most eukaryotic cells, but are not present in conifers (Pinophyta), flowering plants (angiosperms) and most fungi, and are ...
and cilium
The cilium, plural cilia (), is a membrane-bound organelle found on most types of eukaryotic cell, and certain microorganisms known as ciliates. Cilia are absent in bacteria and archaea. The cilium has the shape of a slender threadlike proj ...
, facultatively aerobic mitochondria, sex (meiosis
Meiosis (; , since it is a reductional division) is a special type of cell division of germ cells in sexually-reproducing organisms that produces the gametes, such as sperm or egg cells. It involves two rounds of division that ultimately ...
and syngamy
Fertilisation or fertilization (see spelling differences), also known as generative fertilisation, syngamy and impregnation, is the fusion of gametes to give rise to a new individual organism or offspring and initiate its development. Pro ...
), a dormant cyst with a cell wall of chitin and/or cellulose
Cellulose is an organic compound with the formula , a polysaccharide consisting of a linear chain of several hundred to many thousands of β(1→4) linked D-glucose units. Cellulose is an important structural component of the primary cell wa ...
and peroxisomes. Later endosymbiosis led to the spread of plastids in some lineages.
Although there is still considerable uncertainty in global eukaryote phylogeny, particularly regarding the position of the root, a rough consensus has started to emerge from the phylogenomic studies of the past two decades. The majority of eukaryotes can be placed in one of two large clades dubbed Amorphea (similar in composition to the unikont hypothesis) and the Diaphoretickes
Diaphoretickes () is a major group of eukaryotic organisms, with over 400,000 species. The majority of the earth's biomass that carries out photosynthesis belongs to Diaphoretickes.
Diaphoretickes includes:
* Archaeplastida (comprising red alg ...
, which includes plants and most algal lineages. A third major grouping, the Excavata, has been abandoned as a formal group in the most recent classification of the International Society of Protistologists due to growing uncertainty as to whether its constituent groups belong together. The proposed phylogeny below includes only one group of excavates (Discoba
Excavata is a major supergroup of unicellular organisms belonging to the domain Eukaryota. It was first suggested by Simpson and Patterson in 1999 and introduced by Thomas Cavalier-Smith in 2002 as a formal taxon. It contains a variety of fr ...
), and incorporates the recent proposal that picozoans are close relatives of rhodophytes.
In some analyses, the Hacrobia group ( Haptophyta + Cryptophyta) is placed next to Archaeplastida, but in others it is nested inside the Archaeplastida. However, several recent studies have concluded that Haptophyta and Cryptophyta do not form a monophyletic group. The former could be a sister group to the SAR group, the latter cluster with the Archaeplastida (plants in the broad sense).
The division of the eukaryotes into two primary clades, bikonts ( Archaeplastida + SAR + Excavata) and unikonts
Amorphea are members of a taxonomic supergroup that includes the basal Amoebozoa and Obazoa. That latter contains the Opisthokonta, which includes the Fungi, Animals and the Choanomonada, or Choanoflagellates. The taxonomic affinities of the mem ...
( Amoebozoa + Opisthokonta), derived from an ancestral biflagellar organism and an ancestral uniflagellar organism, respectively, had been suggested earlier. A 2012 study produced a somewhat similar division, although noting that the terms "unikonts" and "bikonts" were not used in the original sense.
A highly converged and congruent set of trees appears in Derelle et al. (2015), Ren et al. (2016), Yang et al. (2017) and Cavalier-Smith (2015) including the supplementary information, resulting in a more conservative and consolidated tree. It is combined with some results from Cavalier-Smith for the basal Opimoda. The main remaining controversies are the root, and the exact positioning of the Rhodophyta and the bikonts Rhizaria, Haptista, Cryptista, Picozoa and Telonemia, many of which may be endosymbiotic eukaryote-eukaryote hybrids. Archaeplastida acquired chloroplast
A chloroplast () is a type of membrane-bound organelle known as a plastid that conducts photosynthesis mostly in plant and algal cells. The photosynthetic pigment chlorophyll captures the energy from sunlight, converts it, and stores it i ...
s probably by endosymbiosis of a prokaryotic ancestor related to a currently extant cyanobacterium, '' Gloeomargarita lithophora''.
Cavalier-Smith's tree
Thomas Cavalier-Smith 2010, 2013, 2014, 2017 and 2018 places the eukaryotic tree's root between Excavata (with ventral feeding groove supported by a microtubular root) and the grooveless Euglenozoa, and monophyletic Chromista, correlated to a single endosymbiotic event of capturing a red-algae. He et al. specifically supports rooting the eukaryotic tree between a monophyleticDiscoba
Excavata is a major supergroup of unicellular organisms belonging to the domain Eukaryota. It was first suggested by Simpson and Patterson in 1999 and introduced by Thomas Cavalier-Smith in 2002 as a formal taxon. It contains a variety of fr ...
(Discicristata
Discicristata is a proposed eukaryotic clade. It consists of Euglenozoa plus Percolozoa.
It was proposed that Discicristata plus Cercozoa yielded Cabozoa. Another proposal is to group Discicristata with Jakobida into Discoba superphylum.
See ...
+ Jakobida) and an Amorphea-Diaphoretickes
Diaphoretickes () is a major group of eukaryotic organisms, with over 400,000 species. The majority of the earth's biomass that carries out photosynthesis belongs to Diaphoretickes.
Diaphoretickes includes:
* Archaeplastida (comprising red alg ...
clade.
Evolutionary history
Origin of eukaryotes

 The origin of the eukaryotic cell, also known as eukaryogenesis, is a milestone in the evolution of life, since eukaryotes include all complex cells and almost all multicellular organisms. A number of approaches have been used to find the first eukaryote and their closest relatives. The last eukaryotic common ancestor (LECA) is the hypothetical last common ancestor of all living eukaryotes, and was most likely a biological population.
Eukaryotes have a number of features that differentiate them from prokaryotes, including an endomembrane system, and unique biochemical pathways such as
The origin of the eukaryotic cell, also known as eukaryogenesis, is a milestone in the evolution of life, since eukaryotes include all complex cells and almost all multicellular organisms. A number of approaches have been used to find the first eukaryote and their closest relatives. The last eukaryotic common ancestor (LECA) is the hypothetical last common ancestor of all living eukaryotes, and was most likely a biological population.
Eukaryotes have a number of features that differentiate them from prokaryotes, including an endomembrane system, and unique biochemical pathways such as sterane
Sterane (cyclopentanoperhydrophenanthrenes) compounds are a class of tetracyclic compounds derived from steroids or sterols via diagenetic and catagenetic degradation and saturation. Steranes have an androstane skeleton with a side chain at carb ...
synthesis. A set of proteins called eukaryotic signature proteins (ESPs) was proposed to identify eukaryotic relatives in 2002: They have no homology to proteins known in other domains of life by then, but they appear to be universal among eukaryotes. They include proteins that make up the cytoskeleton, the complex transcription machinery, membrane-sorting systems, the nuclear pore, as well as some enzymes in the biochemical pathways.
Fossils
The timing of this series of events is hard to determine; Knoll (2006) suggests they developed approximately 1.6–2.1 billion years ago. Some acritarchs are known from at least 1.65 billion years ago, and the possible alga '' Grypania'' has been found as far back as 2.1 billion years ago. The '' Geosiphon''-like fossilfungus
A fungus ( : fungi or funguses) is any member of the group of eukaryotic organisms that includes microorganisms such as yeasts and molds, as well as the more familiar mushrooms. These organisms are classified as a kingdom, separately fr ...
''Diskagma
''Diskagma'' ("disc-like fragment") is a genus of problematic fossil from a Paleoproterozoic (2200 million years old) paleosol from South Africa, and significant as the oldest likely eukaryote and earliest evidence for megascopic life on land.
...
'' has been found in paleosols 2.2 billion years old.
Organized living structures have been found in the black shale
Shale is a fine-grained, clastic sedimentary rock formed from mud that is a mix of flakes of clay minerals (hydrous aluminium phyllosilicates, e.g. kaolin, Al2 Si2 O5( OH)4) and tiny fragments (silt-sized particles) of other minerals, especiall ...
s of the Palaeoproterozoic Francevillian B Formation in Gabon, dated at 2.1 billion years old. Eukaryotic life could have evolved at that time. Fossils that are clearly related to modern groups start appearing an estimated 1.2 billion years ago, in the form of a red algae
Red algae, or Rhodophyta (, ; ), are one of the oldest groups of eukaryotic algae. The Rhodophyta also comprises one of the largest phyla of algae, containing over 7,000 currently recognized species with taxonomic revisions ongoing. The majority ...
, though recent work suggests the existence of fossilized filamentous algae in the Vindhya
The Vindhya Range (also known as Vindhyachal) () is a complex, discontinuous chain of mountain ridges, hill ranges, highlands and plateau escarpments in west-central India.
Technically, the Vindhyas do not form a single mountain range in the ...
basin dating back perhaps to 1.6 to 1.7 billion years ago.
The presence of eukaryotic-specific biomarkers (sterane
Sterane (cyclopentanoperhydrophenanthrenes) compounds are a class of tetracyclic compounds derived from steroids or sterols via diagenetic and catagenetic degradation and saturation. Steranes have an androstane skeleton with a side chain at carb ...
s) in Australia
Australia, officially the Commonwealth of Australia, is a sovereign country comprising the mainland of the Australian continent, the island of Tasmania, and numerous smaller islands. With an area of , Australia is the largest country by ...
n shales previously indicated that eukaryotes were present in these rocks dated at 2.7 billion years old, which was even 300 million years older than the first geological records of the appreciable amount of molecular oxygen during the Great Oxidation Event. However, these Archaean biomarkers were eventually rebutted as later contaminants. Currently, putatively the oldest biomarker records are only ~800 million years old. In contrast, a molecular clock analysis suggests the emergence of sterol biosynthesis as early as 2.3 billion years ago, and thus there is a huge gap between molecular data and geological data, which hinders a reasonable inference of the eukaryotic evolution through biomarker records before 800 million years ago. The nature of steranes as eukaryotic biomarkers is further complicated by the production of sterols by some bacteria.
Whenever their origins, eukaryotes may not have become ecologically dominant until much later; a massive uptick in the zinc composition of marine sediments has been attributed to the rise of substantial populations of eukaryotes, which preferentially consume and incorporate zinc relative to prokaryotes, approximately a billion years after their origin (at the latest).
In April 2019, biologists reported that the very large medusavirus
Medusavirus is a nucleocytoplasmic large DNA virus first isolated from a Japanese hot spring in 2019. It notably encodes all five types of histones — H1, H2A, H2B, H3, and H4 — which are involved in DNA packaging in eukaryotes, raising the p ...
, or a relative, may have been responsible, at least in part, for the evolutionary emergence of complex eukaryotic cells from simpler prokaryotic cells.
*
Relationship to Archaea
The nuclear DNA and genetic machinery of eukaryotes is more similar to Archaea thanBacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were am ...
, leading to a controversial suggestion that eukaryotes should be grouped with Archaea in the clade Neomura
Neomura is a possible clade composed of the two domains of life of Archaea and Eukaryota. The group was named by Thomas Cavalier-Smith in 2002. Its name means "new walls", reflecting his hypothesis that it evolved from Bacteria, and one of t ...
. In other respects, such as membrane composition, eukaryotes are similar to Bacteria. Three main explanations for this have been proposed:
* Eukaryotes resulted from the complete fusion of two or more cells, wherein the cytoplasm formed from a bacterium
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were am ...
, and the nucleus from an archaeon, from a virus, or from a pre-cell A pre-cell (or protocell) is a hypothetical lipid-based structure that, under the RNA world hypothesis, could have confined RNA in ancient times. A pre-cell allowed the RNA to remain in close proximity with other RNA molecules, keeping them concentr ...
.
* Eukaryotes developed from Archaea, and acquired their bacterial characteristics through the endosymbiosis of a proto-mitochondrion
The proto-mitochondrion is the hypothetical ancestral bacterial endosymbiont from which all mitochondria in eukaryotes are thought to descend, after an episode of symbiogenesis which created the aerobic eukaryotes.
Phylogeny
The phylogenetic ana ...
of bacterial origin.
* Eukaryotes and Archaea developed separately from a modified bacterium.
rRNA
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribos ...
analysis supports the eocyte scenario, apparently with the Eukaryote root in Excavata. A cladogram supporting the eocyte hypothesis, positioning eukaryotes within Archaea, based on phylogenomic analyses of the Asgard archaea, is:
In this scenario, the Asgard group is seen as a sister taxon of the TACK
TACK is a group of archaea acronym for Thaumarchaeota (now Nitrososphaerota), Aigarchaeota, Crenarchaeota (now Thermoproteota), and Korarchaeota, the first groups discovered. They are found in different environments ranging from acidophilic ...
group, which comprises Thermoproteota (formerly named eocytes or Crenarchaeota), Nitrososphaerota (formerly Thaumarchaeota), and others. This group is reported contain many of the eukaryotic signature proteins and produce vesicles.
In 2017, there was significant pushback against this scenario, arguing that the eukaryotes did not emerge within the Archaea. Cunha ''et al.'' produced analyses supporting the three domains (3D) or Woese hypothesis (2 in the table above) and rejecting the eocyte hypothesis (4 above). Harish and Kurland found strong support for the earlier two empires (2D) or Mayr hypothesis (1 in the table above), based on analyses of the coding sequences of protein domains. They rejected the eocyte hypothesis as the least likely. A possible interpretation of their analysis is that the universal common ancestor (UCA) of the current tree of life was a complex organism that survived an evolutionary bottleneck
A population bottleneck or genetic bottleneck is a sharp reduction in the size of a population due to environmental events such as famines, earthquakes, floods, fires, disease, and droughts; or human activities such as specicide, widespread violen ...
, rather than a simpler organism arising early in the history of life. On the other hand, the researchers who came up with Asgard re-affirmed their hypothesis with additional Asgard samples. Since then, the publication of additional Asgard archaeal genomes and the independent reconstruction of phylogenomic trees by multiple independent laboratories have provided additional support for an Asgard archaeal origin of eukaryotes.
Details of the relation of Asgard archaea members and eukaryotes are still under consideration, although, in January 2020, scientists reported that ''Candidatus Prometheoarchaeum syntrophicum
Lokiarchaeota is a proposed phylum of the Archaea. The phylum includes all members of the group previously named Deep Sea Archaeal Group (DSAG), also known as Marine Benthic Group B (MBG-B). Lokiarchaeota is part of the superphylum Asgard contai ...
'', a type of cultured Asgard archaea, may be a possible link between simple prokaryotic and complex eukaryotic
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacte ...
microorganisms about two billion years ago.
Endomembrane system and mitochondria
The origins of the endomembrane system and mitochondria are also unclear. The phagotrophic hypothesis proposes that eukaryotic-type membranes lacking a cell wall originated first, with the development of endocytosis, whereas mitochondria were acquired by ingestion as endosymbionts. The syntrophic hypothesis proposes that the proto-eukaryote relied on the proto-mitochondrion for food, and so ultimately grew to surround it. Here the membranes originated after the engulfment of the mitochondrion, in part thanks to mitochondrial genes (thehydrogen hypothesis The hydrogen hypothesis is a model proposed by William F. Martin and Miklós Müller in 1998 that describes a possible way in which the mitochondrion arose as an endosymbiont within a prokaryotic host in the archaea, giving rise to a symbiotic asso ...
is one particular version).
In a study using genomes to construct supertree A supertree is a single phylogenetic tree assembled from a combination of smaller phylogenetic trees, which may have been assembled using different datasets (e.g. morphological and molecular) or a different selection of taxa. Supertree algorithms c ...
s, Pisani ''et al.'' (2007) suggest that, along with evidence that there was never a mitochondrion-less eukaryote, eukaryotes evolved from a syntrophy between an archaea closely related to Thermoplasmatales and an alphaproteobacterium
Alphaproteobacteria is a class of bacteria in the phylum Pseudomonadota (formerly Proteobacteria). The Magnetococcales and Mariprofundales are considered basal or sister to the Alphaproteobacteria. The Alphaproteobacteria are highly diverse an ...
, likely a symbiosis driven by sulfur or hydrogen. The mitochondrion and its genome is a remnant of the alphaproteobacterial endosymbiont. The majority of the genes from the symbiont have been transferred to the nucleus. They make up most of the metabolic and energy-related pathways of the eukaryotic cell, while the information system (DNA polymerase, transcription, translation) is retained from archaea.
Hypotheses
Different hypotheses have been proposed as to how eukaryotic cells came into existence. These hypotheses can be classified into two distinct classes – autogenous models and chimeric models.=Autogenous models
= Autogenous models propose that a proto-eukaryotic cell containing anucleus
Nucleus ( : nuclei) is a Latin word for the seed inside a fruit. It most often refers to:
* Atomic nucleus, the very dense central region of an atom
*Cell nucleus, a central organelle of a eukaryotic cell, containing most of the cell's DNA
Nucl ...
existed first, and later acquired mitochondria
A mitochondrion (; ) is an organelle found in the cells of most Eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used ...
. According to this model, a large prokaryote
A prokaryote () is a single-celled organism that lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Greek πρό (, 'before') and κάρυον (, 'nut' or 'kernel').Campbell, N. "Biology:Concepts & Con ...
developed invaginations
Invagination is the process of a surface folding in on itself to form a cavity, pouch or tube. In developmental biology, invagination is a mechanism that takes place during gastrulation. This mechanism or cell movement happens mostly in the vegetal ...
in its plasma membrane in order to obtain enough surface area
The surface area of a solid object is a measure of the total area that the surface of the object occupies. The mathematical definition of surface area in the presence of curved surfaces is considerably more involved than the definition of ...
to service its cytoplasmic volume. As the invaginations differentiated in function, some became separate compartments – giving rise to the endomembrane system, including the endoplasmic reticulum
The endoplasmic reticulum (ER) is, in essence, the transportation system of the eukaryotic cell, and has many other important functions such as protein folding. It is a type of organelle made up of two subunits – rough endoplasmic reticulum ...
, golgi apparatus
The Golgi apparatus (), also known as the Golgi complex, Golgi body, or simply the Golgi, is an organelle found in most eukaryotic cells. Part of the endomembrane system in the cytoplasm, it packages proteins into membrane-bound vesicles i ...
, nuclear membrane, and single membrane structures such as lysosome
A lysosome () is a membrane-bound organelle found in many animal cells. They are spherical vesicles that contain hydrolytic enzymes that can break down many kinds of biomolecules. A lysosome has a specific composition, of both its membrane p ...
s.
Mitochondria
A mitochondrion (; ) is an organelle found in the cells of most Eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used ...
are proposed to come from the endosymbiosis of an aerobic proteobacterium
Pseudomonadota (synonym Proteobacteria) is a major phylum of Gram-negative bacteria. The renaming of phyla in 2021 remains controversial among microbiologists, many of whom continue to use the earlier names of long standing in the literature. The ...
after an eukaryote with a nucleus has evolved. This theory is less held onto because it requires extra assumptions to explain current conditions. For example, as every known eukaryote has a mitochondrion (or at least show signs of having an ancestor that had), one must assumed that all the eukaryotic lineages that did not acquire mitochondria became extinct. The theory also doesn’t explain why anaerobic variants of mitochondria have evolved.
=Chimeric models
= Chimeric models claim that two prokaryotic cells existed initially – an archaeon and abacterium
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were am ...
. The closest living relatives of these appears to be Asgardarchaeota and (distantly related) the alphaproteobacteria called the proto-mitochondrion
The proto-mitochondrion is the hypothetical ancestral bacterial endosymbiont from which all mitochondria in eukaryotes are thought to descend, after an episode of symbiogenesis which created the aerobic eukaryotes.
Phylogeny
The phylogenetic ana ...
.
* These cells underwent a merging process, either by a physical fusion or by endosymbiosis, thereby leading to the formation of a eukaryotic cell. Within these chimeric models, some studies further claim that mitochondria originated from a bacterial ancestor while others emphasize the role of endosymbiotic processes behind the origin of mitochondria.
=The inside-out hypothesis
= The inside-out hypothesis suggests that the fusion between free-living mitochondria-like bacteria, and an archaeon into a eukaryotic cell happened gradually over a long period of time, instead of in a single phagocytotic event. In this scenario, an archaeon would trap aerobic bacteria with cell protrusions, and then keep them alive to draw energy from them instead of digesting them. During the early stages the bacteria would still be partly in direct contact with the environment, and the archaeon would not have to provide them with all the required nutrients. But eventually the archaeon would engulf the bacteria completely, creating the internal membrane structures and nucleus membrane in the process. * It is assumed the archaean group called halophiles went through a similar procedure, where they acquired as much as a thousand genes from a bacterium, way more than through the conventional horizontal gene transfer that often occurs in the microbial world, but that the two microbes separated again before they had fused into a single eukaryote-like cell. An expanded version of the inside-out hypothesis proposes that the eukaryotic cell was created by physical interactions between two prokaryotic organisms and that the last common ancestor of eukaryotes got its genome from a whole population or community of microbes participating in cooperative relationships to thrive and survive in their environment. The genome from the various types of microbes would complement each other, and occasional horizontal gene transfer between them would be largely to their own benefit. This accumulation of beneficial genes gave rise to the genome of the eukaryotic cell, which contained all the genes required for independence.=The serial endosymbiotic hypothesis
= According to serial endosymbiotic theory (championed by Lynn Margulis), a union between a motile anaerobic bacterium (like ''Spirochaeta'') and a thermoacidophiliccrenarchaeon
The Thermoproteota (also known as crenarchaea) are archaea that have been classified as a phylum of the Archaea domain. Initially, the Thermoproteota were thought to be sulfur-dependent extremophiles but recent studies have identified characteri ...
(like ''Thermoplasma'' which is sulfidogenic in nature) gave rise to the present day eukaryotes. This union established a motile organism capable of living in the already existing acidic and sulfurous waters. Oxygen is known to cause toxicity to organisms that lack the required metabolic machinery. Thus, the archaeon provided the bacterium with a highly beneficial reduced environment (sulfur and sulfate were reduced to sulfide). In microaerophilic conditions, oxygen was reduced to water thereby creating a mutual benefit platform. The bacterium on the other hand, contributed the necessary fermentation products and electron
The electron ( or ) is a subatomic particle with a negative one elementary electric charge. Electrons belong to the first generation of the lepton particle family,
and are generally thought to be elementary particles because they have n ...
acceptors along with its motility feature to the archaeon thereby gaining a swimming motility
Motility is the ability of an organism to move independently, using metabolic energy.
Definitions
Motility, the ability of an organism to move independently, using metabolic energy, can be contrasted with sessility, the state of organisms th ...
for the organism.
From a consortium of bacterial and archaeal DNA originated the nuclear genome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding ...
of eukaryotic cells. Spirochetes gave rise to the motile features of eukaryotic cells. Endosymbiotic unifications of the ancestors of alphaproteobacteria and cyanobacteria, led to the origin of mitochondria and plastids respectively. For example, ''Thiodendron'' has been known to have originated via an ectosymbiotic process based on a similar syntrophy of sulfur existing between the two types of bacteria – ''Desulfobacter
''Desulfobacter'' is a genus of bacteria from the family of Desulfobacteraceae. ''Desulfobacter'' has the ability to oxidize acetate
An acetate is a salt formed by the combination of acetic acid with a base (e.g. alkaline, earthy, metal ...
'' and ''Spirochaeta''.
However, such an association based on motile symbiosis has never been observed practically. Also there is no evidence of archaeans and spirochetes adapting to intense acid-based environments. In addition, the theory posits that mitochondrion-less eukaryotes have existed, tying back to the problem in the autogenous model.
=The hydrogen hypothesis
= In thehydrogen hypothesis The hydrogen hypothesis is a model proposed by William F. Martin and Miklós Müller in 1998 that describes a possible way in which the mitochondrion arose as an endosymbiont within a prokaryotic host in the archaea, giving rise to a symbiotic asso ...
, the symbiotic linkage of an anaerobic and autotrophic methanogenic archaeon (host) with an alphaproteobacterium (the symbiont) gave rise to the eukaryotes. The host used hydrogen
Hydrogen is the chemical element with the symbol H and atomic number 1. Hydrogen is the lightest element. At standard conditions hydrogen is a gas of diatomic molecules having the formula . It is colorless, odorless, tasteless, non-to ...
(H2) and carbon dioxide
Carbon dioxide ( chemical formula ) is a chemical compound made up of molecules that each have one carbon atom covalently double bonded to two oxygen atoms. It is found in the gas state at room temperature. In the air, carbon dioxide is t ...
() to produce methane
Methane ( , ) is a chemical compound with the chemical formula (one carbon atom bonded to four hydrogen atoms). It is a group-14 hydride, the simplest alkane, and the main constituent of natural gas. The relative abundance of methane ...
while the symbiont, capable of aerobic respiration, expelled H2 and as byproducts of anaerobic fermentation process. The host's methanogenic environment worked as a sink for H2, which resulted in heightened bacterial fermentation.
Endosymbiotic gene transfer
A diatom ( Neo-Latin ''diatoma''), "a cutting through, a severance", from el, διάτομος, diátomos, "cut in half, divided equally" from el, διατέμνω, diatémno, "to cut in twain". is any member of a large group comprising se ...
acted as a catalyst for the host to acquire the symbionts' carbohydrate
In organic chemistry, a carbohydrate () is a biomolecule consisting of carbon (C), hydrogen (H) and oxygen (O) atoms, usually with a hydrogen–oxygen atom ratio of 2:1 (as in water) and thus with the empirical formula (where ''m'' may o ...
metabolism and turn heterotroph
A heterotroph (; ) is an organism that cannot produce its own food, instead taking nutrition from other sources of organic carbon, mainly plant or animal matter. In the food chain, heterotrophs are primary, secondary and tertiary consumers, but ...
ic in nature. Subsequently, the host's methane forming capability was lost. Thus, the origins of the heterotrophic organelle (symbiont) are identical to the origins of the eukaryotic lineage
Lineage may refer to:
Science
* Lineage (anthropology), a group that can demonstrate its common descent from an apical ancestor or a direct line of descent from an ancestor
* Lineage (evolution), a temporal sequence of individuals, populat ...
. In this hypothesis, the presence of H2 represents the selective force that forged eukaryotes out of prokaryotes.
=The syntrophy hypothesis
= The syntrophy hypothesis was developed in contrast to the hydrogen hypothesis and proposes the existence of two symbiotic events. According to this model, the origin of eukaryotic cells was based on metabolic symbiosis (syntrophy) between a methanogenic archaeon and adeltaproteobacterium
The Myxococcota are a phylum of bacteria known as the fruiting gliding bacteria. All species of this group are Gram-negative. They are predominantly aerobic genera that release myxospores in unfavorable environments.
Phylogeny
The currently ac ...
. This syntrophic symbiosis was initially facilitated by H2 transfer between different species under anaerobic environments. In earlier stages, an alphaproteobacterium became a member of this integration, and later developed into the mitochondrion. Gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b ...
transfer from a deltaproteobacterium to an archaeon led to the methanogenic archaeon developing into a nucleus. The archaeon constituted the genetic apparatus, while the deltaproteobacterium contributed towards the cytoplasm
In cell biology, the cytoplasm is all of the material within a eukaryotic cell, enclosed by the cell membrane, except for the cell nucleus. The material inside the nucleus and contained within the nuclear membrane is termed the nucleoplasm. ...
ic features.
This theory incorporates two selective forces at the time of nucleus evolution
Evolution is change in the heritable characteristics of biological populations over successive generations. These characteristics are the expressions of genes, which are passed on from parent to offspring during reproduction. Variation ...
* presence of metabolic partitioning to avoid the harmful effects of the co-existence of anabolic
Anabolism () is the set of metabolic pathways that construct molecules from smaller units. These reactions require energy, known also as an endergonic process. Anabolism is the building-up aspect of metabolism, whereas catabolism is the breakin ...
and catabolic
Catabolism () is the set of metabolic pathways that breaks down molecules into smaller units that are either oxidized to release energy or used in other anabolic reactions. Catabolism breaks down large molecules (such as polysaccharides, lip ...
cellular pathways, and
* prevention of abnormal protein biosynthesis due to a vast spread of introns in the archaeal genes after acquiring the mitochondrion and losing methanogenesis.
=6+ serial endosymbiosis scenario
= A complex scenario of 6+ serial endosymbiotic events of archaea and bacteria has been proposed in which mitochondria and an asgard related archaeota were acquired at a late stage of eukaryogenesis, possibly in combination, as a secondary endosymbiont. The findings have been rebuked as an artifact.See also
* Eukaryote hybrid genome * Evolution of sexual reproduction * List of sequenced eukaryotic genomes * ''Parakaryon myojinensis
''Parakaryon myojinensis'', also known as the Myojin parakaryote, is a highly unusual species of single-celled organism known only from a single specimen, described in 2012. It has features of both prokaryotes and eukaryotes but is apparently d ...
''
* Prokaryote
A prokaryote () is a single-celled organism that lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Greek πρό (, 'before') and κάρυον (, 'nut' or 'kernel').Campbell, N. "Biology:Concepts & Con ...
* Nitrososphaerota
* Vault (organelle)
The vault or vault cytoplasmic ribonucleoprotein is a eukaryotic organelle whose function is not yet fully understood. Discovered and isolated by Nancy Kedersha and Leonard Rome in 1986, vaults are cytoplasmic organelles which, when negative- ...
Notes
References
External links
"Eukaryotes"
( Tree of Life Web Project) *
Attraction and sex among our microbial Last Eukaryotic Common Ancestors
The Atlantic, November 11, 2020 {{Authority control Articles containing video clips Eukaryote